Pre Gene Modal
BGIBMGA012163
Annotation
PREDICTED:_U4/U6_small_nuclear_ribonucleoprotein_Prp3_isoform_X1_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 2.952
Sequence
CDS
ATGGCACTTCAGCTGTCCCGTCGCGAGGTAGAGGAACTCCGGTCATCAATAGACCGTGCCATATACAGGACTATAGGAAAATCGGATAACTCAATTCTATCGACTGTTTCGTCGTGTTTGACGAACGGATATGAACGTAGAAAAATTATTGACCGACTTTCATCGCACATAGACTCCAAAAAGGCAAGCAAACTAACCGATAAGGTCATAGCATTGGCCCAGGAACTGATAACGTCATCAAAAACTTCATCGAAAAGGAAACATGAGAGTGACAGAGATAGAGATGGTAAGAGATCTCGGTACGAAGGTTCAAAGCAAGATGAGAAAGTTTATGTGGAGAAAAAAGATGTGGAGGAAAAGAATGGAACTGATGATCTTCCCGCTATTCCCGAAGGAGAGACTCCAGGATCGAAGATGGCTATCCTGAGTGCAGACAGGATTAAGTTAATGATGGCCAACGCTCAGAAAGAGATAGAGGAACGCAAGAGAGCTCTTCAAGTGCTGAAGGGGGTTGAGACCAAACCGAGTGTTAATGCTGCGGTGGCTGCGGCCCAACACCTGAAAGTGCAGATGCAGCAGAACAATGCAATACCCCCACCGAGTGTAATCAAACCTGTACTGTATTCAAAACCTGAAACTTTGCCGTTATCACGCGAAGAATTGGAGAAGCAAAGGAAGATAGCTGAGCTGCAAGCGAGGATACAGCGGAAACTTGCAGGGGGTGCCTTATCATCGACGCCGGGGTCGGGACCTGCCCCCCTGATCTTGGACCGTGAGGGTCGCACCGTGGACACCACGGGCAGGATAGTTCAACTCACCCACGTGGCGCCCACGCTTCAGGCGAACATAAGAGCGAAGAGACGTGAGGAGTTCAAAGCTCAGTTGTCCGGTCCCACTGAGGAGGTGGGTCCGGAGTCGTCCCGCTGGGTGGACCCGCGCGTGTCCTCGCGCCCCCACGCGCGCGCCAAGCGGCCCCTGCGCTTCCACGAGCCCGGCAAGTTCCGCCAGATGGCTGAACGGCTAAGGATGAAGGCGCAACTGGAGAAGCTGCAGAACGAGATATCCCAGATAGCGCGGAAGACCGGCATCTCGTCCGCCACCAGGTTAGCGCTGCTGGCGGCCGACGCGCCCGACGCCGACGGACCCCCCGACGTGGAATGGTGGGACAGCGTGATCCTGATGACGGCGGACGAGCGGGAGACGAAGCGGCGCGGGGCCGGCGCTGATGGGCGCACGGACGAGCAGCGCGTGGACGCCTGCAACACGGGCGAGGACGACATAGTGGACAACATCAACGAGGAGGCCATCACCAACCTCGTGGAGCACCCGCAGCAGCTCAGGCCGCCCACGGAACCTCTCAAGCCGACGTTCATGCCGGTCTTCCTGACGAAGAAGGAGAGGAAGAAACTGCGACGCCAGAGCAGACGGGAGGCTTGGAAGGAGGAGCAGGAGAAGGTCAGACTCGGACTGGAAGCGCCACCAGAACCAAAGCTCAGGATCTCGAATCTGATGCGAGCTTTAGGCACCGAAGCGGTGCAGGACCCCACGGCGATCGAGGCCAAGGTCAGGGAGCAGATTGCGAAGAGACAGAAGACGCATCTGGAAGCGAACAAGGCGAGAGCTCTCACCAAGGAACAGAAGAGGGAGAAGGTTGACCGCAAGCTGCAGGAAGACACGTCGGCCGGCGTCCACGTGGCCGTGTACAGGATCCGTGATCTGTTCGAGTGCGCGTCGGCCAAGTTCAAGGTCGAGGTGAACGCCCAGCAGCTGCACATGAGCGGCTGCGTGCTGCTGCACCGGGGCTGCTGCGTGGTCGTGGCCGAGGGGGGCCCCCGCCAGCACGCCAAGTACAAGCGGTTGATGCTGCACCGCATTAAGTGGGAGGAGGACACCGTTAAGGACGCCGAAGGCTGCGACGTGCCCAACTCCTGTCACCTGGTCTGGGAGGGCACCACCAAACAAAGGGCCTTCGGGGAGATCAAGTTTAAGGTGGTACCGACAGAGAAACAAGCGCGCGAGCATTTCCAAAAACACGGCGTCGAACATTACTGGGACCTGGCGTACAGCAGTGCGGTCCTCGGACCCACTATAGAGGAGCCGTGA
Protein
MALQLSRREVEELRSSIDRAIYRTIGKSDNSILSTVSSCLTNGYERRKIIDRLSSHIDSKKASKLTDKVIALAQELITSSKTSSKRKHESDRDRDGKRSRYEGSKQDEKVYVEKKDVEEKNGTDDLPAIPEGETPGSKMAILSADRIKLMMANAQKEIEERKRALQVLKGVETKPSVNAAVAAAQHLKVQMQQNNAIPPPSVIKPVLYSKPETLPLSREELEKQRKIAELQARIQRKLAGGALSSTPGSGPAPLILDREGRTVDTTGRIVQLTHVAPTLQANIRAKRREEFKAQLSGPTEEVGPESSRWVDPRVSSRPHARAKRPLRFHEPGKFRQMAERLRMKAQLEKLQNEISQIARKTGISSATRLALLAADAPDADGPPDVEWWDSVILMTADERETKRRGAGADGRTDEQRVDACNTGEDDIVDNINEEAITNLVEHPQQLRPPTEPLKPTFMPVFLTKKERKKLRRQSRREAWKEEQEKVRLGLEAPPEPKLRISNLMRALGTEAVQDPTAIEAKVREQIAKRQKTHLEANKARALTKEQKREKVDRKLQEDTSAGVHVAVYRIRDLFECASAKFKVEVNAQQLHMSGCVLLHRGCCVVVAEGGPRQHAKYKRLMLHRIKWEEDTVKDAEGCDVPNSCHLVWEGTTKQRAFGEIKFKVVPTEKQAREHFQKHGVEHYWDLAYSSAVLGPTIEEP
Summary
Uniprot
H9JRK3
A0A0N0PA47
A0A212EYQ2
A0A0T6B316
A0A1W4W5F5
A0A1Y1KF46
+ More
D6WBT3 A0A154PLF8 A0A067RKW1 A0A2A3EKW9 A0A087ZTY5 E2AB89 A0A0J7KV41 A0A310SWP3 A0A2J7Q6E0 Q16P69 A0A0N0U5W3 A0A158NYM0 A0A026WCF1 A0A195DEI9 A0A151WK86 A0A195CFF8 A0A182GW90 B0WFV6 A0A0L7RI44 A0A0C9PU88 E2B9D0 T1PI01 A0A1Q3EWJ2 A0A0A1WJF6 W8BRE3 A0A034WGB4 K7J6K3 A0A195BS51 A0A034WFA1 A0A232F0H9 A0A1I8NWM7 A0A1B0A6A0 A0A1A9Y2T9 A0A2M4BFL8 A0A1B0AWD4 A0A1B0FEZ3 A0A1A9VKW5 A0A0L0BNG6 W5JX69 A0A2M4APY7 A0A1Y1KMQ2 A0A1I8M8Y6 A0A1A9WSR4 F4W9P9 A0A195FGR7 A0A1B0CKP2 A0A2S2QBA2 A0A0A1XA67 A0A1I8NWN0 A0A224XG85 W8BX98 A0A023F2J9 A0A2H8TLC1 A0A182IZR2 B4ML95 B4IIS2 B4L0W5 B4LBL7 B3M8T4 A0A0Q9WPC4 A0A0Q9XR12 B4QRC8 Q9VW52 A0A1W4W6B9 A0A0Q9WU09 B3NE03 B4PFW5 A0A182YDW1 A0A084VJH2 Q2LYV5 B4H035 A0A0A9W7U3 A0A3B0J5J8 N6UNA6 A0A1L8DGI3 N6TQT2 A0A1J1HE94 A0A0J9RYA9 A0A0P8XW31 Q8MRG6 B4IXN8 A0A1W4VTR7 A0A0Q5UGC8 A0A0R1E0P9 A0A182W7Z7 A0A0M3QWS9 A0A182N8D3 N6UQ17 T1I1M9 A0A182JR22 A0A069DV86 V5IG47 T1J0D7
D6WBT3 A0A154PLF8 A0A067RKW1 A0A2A3EKW9 A0A087ZTY5 E2AB89 A0A0J7KV41 A0A310SWP3 A0A2J7Q6E0 Q16P69 A0A0N0U5W3 A0A158NYM0 A0A026WCF1 A0A195DEI9 A0A151WK86 A0A195CFF8 A0A182GW90 B0WFV6 A0A0L7RI44 A0A0C9PU88 E2B9D0 T1PI01 A0A1Q3EWJ2 A0A0A1WJF6 W8BRE3 A0A034WGB4 K7J6K3 A0A195BS51 A0A034WFA1 A0A232F0H9 A0A1I8NWM7 A0A1B0A6A0 A0A1A9Y2T9 A0A2M4BFL8 A0A1B0AWD4 A0A1B0FEZ3 A0A1A9VKW5 A0A0L0BNG6 W5JX69 A0A2M4APY7 A0A1Y1KMQ2 A0A1I8M8Y6 A0A1A9WSR4 F4W9P9 A0A195FGR7 A0A1B0CKP2 A0A2S2QBA2 A0A0A1XA67 A0A1I8NWN0 A0A224XG85 W8BX98 A0A023F2J9 A0A2H8TLC1 A0A182IZR2 B4ML95 B4IIS2 B4L0W5 B4LBL7 B3M8T4 A0A0Q9WPC4 A0A0Q9XR12 B4QRC8 Q9VW52 A0A1W4W6B9 A0A0Q9WU09 B3NE03 B4PFW5 A0A182YDW1 A0A084VJH2 Q2LYV5 B4H035 A0A0A9W7U3 A0A3B0J5J8 N6UNA6 A0A1L8DGI3 N6TQT2 A0A1J1HE94 A0A0J9RYA9 A0A0P8XW31 Q8MRG6 B4IXN8 A0A1W4VTR7 A0A0Q5UGC8 A0A0R1E0P9 A0A182W7Z7 A0A0M3QWS9 A0A182N8D3 N6UQ17 T1I1M9 A0A182JR22 A0A069DV86 V5IG47 T1J0D7
Pubmed
19121390
26354079
22118469
28004739
18362917
19820115
+ More
24845553 20798317 17510324 21347285 24508170 26483478 25315136 25830018 24495485 25348373 20075255 28648823 26108605 20920257 23761445 21719571 25474469 17994087 18057021 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 25244985 24438588 15632085 25401762 26823975 23537049 26109357 26109356 26334808 25765539
24845553 20798317 17510324 21347285 24508170 26483478 25315136 25830018 24495485 25348373 20075255 28648823 26108605 20920257 23761445 21719571 25474469 17994087 18057021 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 25244985 24438588 15632085 25401762 26823975 23537049 26109357 26109356 26334808 25765539
EMBL
BABH01031780
BABH01031781
BABH01031782
BABH01031783
BABH01031784
BABH01031785
+ More
KQ458863 KPJ04715.1 AGBW02011451 OWR46626.1 LJIG01016025 KRT81839.1 GEZM01085211 GEZM01085207 JAV60122.1 KQ971314 EEZ97870.1 KQ434968 KZC12705.1 KK852411 KDR24472.1 KZ288227 PBC31836.1 GL438234 EFN69365.1 LBMM01002796 KMQ94352.1 KQ759849 OAD62566.1 NEVH01017533 PNF24142.1 CH477792 EAT36165.1 KQ435752 KOX76182.1 ADTU01000683 ADTU01000684 KK107321 EZA52724.1 KQ980979 KYN10839.1 KQ983012 KYQ48289.1 KQ977876 KYM99051.1 JXUM01092887 KQ564017 KXJ72933.1 DS231921 EDS26491.1 KQ414588 KOC70494.1 GBYB01004918 JAG74685.1 GL446499 EFN87681.1 KA648442 AFP63071.1 GFDL01015358 JAV19687.1 GBXI01015647 JAC98644.1 GAMC01002660 JAC03896.1 GAKP01006144 JAC52808.1 AAZX01001310 KQ976423 KYM88832.1 GAKP01006142 JAC52810.1 NNAY01001410 OXU24069.1 GGFJ01002699 MBW51840.1 JXJN01004653 CCAG010017197 JRES01001600 KNC21577.1 ADMH02000009 ETN68110.1 GGFK01009534 MBW42855.1 GEZM01085210 JAV60117.1 GL888033 EGI69035.1 KQ981606 KYN39432.1 AJWK01016490 GGMS01005803 MBY75006.1 GBXI01006098 JAD08194.1 GFTR01007598 JAW08828.1 GAMC01002658 JAC03898.1 GBBI01002870 JAC15842.1 GFXV01003142 MBW14947.1 AXCP01007317 CH963847 EDW73353.1 CH480844 EDW49843.1 CH933809 EDW19215.1 KRG06551.1 CH940647 EDW70827.1 CH902618 EDV41085.1 KRF97749.1 KRG06552.1 CM000363 CM002912 EDX11141.1 KMZ00638.1 AE014296 AAF49097.1 KRF85137.1 CH954178 EDV52427.1 CM000159 EDW95262.1 ATLV01013567 KE524868 KFB38116.1 CH379069 EAL29755.1 CH479199 EDW29612.1 GBHO01040118 GBHO01013534 GBRD01005039 GDHC01015711 GDHC01007554 JAG03486.1 JAG30070.1 JAG60782.1 JAQ02918.1 JAQ11075.1 OUUW01000002 SPP77264.1 APGK01009162 APGK01024977 KB740570 KB737872 KB632375 ENN80207.1 ENN82862.1 ERL93888.1 GFDF01008587 JAV05497.1 APGK01009169 KB737873 ENN82859.1 CVRI01000001 CRK86288.1 KMZ00639.1 KPU78915.1 AY119640 AAM50294.1 AAN11640.1 CH916366 EDV97500.1 KQS44273.1 KRK02334.1 CP012525 ALC44639.1 ENN82861.1 ACPB03016272 GBGD01000911 JAC87978.1 GANP01007317 JAB77151.1 JH431734
KQ458863 KPJ04715.1 AGBW02011451 OWR46626.1 LJIG01016025 KRT81839.1 GEZM01085211 GEZM01085207 JAV60122.1 KQ971314 EEZ97870.1 KQ434968 KZC12705.1 KK852411 KDR24472.1 KZ288227 PBC31836.1 GL438234 EFN69365.1 LBMM01002796 KMQ94352.1 KQ759849 OAD62566.1 NEVH01017533 PNF24142.1 CH477792 EAT36165.1 KQ435752 KOX76182.1 ADTU01000683 ADTU01000684 KK107321 EZA52724.1 KQ980979 KYN10839.1 KQ983012 KYQ48289.1 KQ977876 KYM99051.1 JXUM01092887 KQ564017 KXJ72933.1 DS231921 EDS26491.1 KQ414588 KOC70494.1 GBYB01004918 JAG74685.1 GL446499 EFN87681.1 KA648442 AFP63071.1 GFDL01015358 JAV19687.1 GBXI01015647 JAC98644.1 GAMC01002660 JAC03896.1 GAKP01006144 JAC52808.1 AAZX01001310 KQ976423 KYM88832.1 GAKP01006142 JAC52810.1 NNAY01001410 OXU24069.1 GGFJ01002699 MBW51840.1 JXJN01004653 CCAG010017197 JRES01001600 KNC21577.1 ADMH02000009 ETN68110.1 GGFK01009534 MBW42855.1 GEZM01085210 JAV60117.1 GL888033 EGI69035.1 KQ981606 KYN39432.1 AJWK01016490 GGMS01005803 MBY75006.1 GBXI01006098 JAD08194.1 GFTR01007598 JAW08828.1 GAMC01002658 JAC03898.1 GBBI01002870 JAC15842.1 GFXV01003142 MBW14947.1 AXCP01007317 CH963847 EDW73353.1 CH480844 EDW49843.1 CH933809 EDW19215.1 KRG06551.1 CH940647 EDW70827.1 CH902618 EDV41085.1 KRF97749.1 KRG06552.1 CM000363 CM002912 EDX11141.1 KMZ00638.1 AE014296 AAF49097.1 KRF85137.1 CH954178 EDV52427.1 CM000159 EDW95262.1 ATLV01013567 KE524868 KFB38116.1 CH379069 EAL29755.1 CH479199 EDW29612.1 GBHO01040118 GBHO01013534 GBRD01005039 GDHC01015711 GDHC01007554 JAG03486.1 JAG30070.1 JAG60782.1 JAQ02918.1 JAQ11075.1 OUUW01000002 SPP77264.1 APGK01009162 APGK01024977 KB740570 KB737872 KB632375 ENN80207.1 ENN82862.1 ERL93888.1 GFDF01008587 JAV05497.1 APGK01009169 KB737873 ENN82859.1 CVRI01000001 CRK86288.1 KMZ00639.1 KPU78915.1 AY119640 AAM50294.1 AAN11640.1 CH916366 EDV97500.1 KQS44273.1 KRK02334.1 CP012525 ALC44639.1 ENN82861.1 ACPB03016272 GBGD01000911 JAC87978.1 GANP01007317 JAB77151.1 JH431734
Proteomes
UP000005204
UP000053268
UP000007151
UP000192223
UP000007266
UP000076502
+ More
UP000027135 UP000242457 UP000005203 UP000000311 UP000036403 UP000235965 UP000008820 UP000053105 UP000005205 UP000053097 UP000078492 UP000075809 UP000078542 UP000069940 UP000249989 UP000002320 UP000053825 UP000008237 UP000095301 UP000002358 UP000078540 UP000215335 UP000095300 UP000092445 UP000092443 UP000092460 UP000092444 UP000078200 UP000037069 UP000000673 UP000091820 UP000007755 UP000078541 UP000092461 UP000075880 UP000007798 UP000001292 UP000009192 UP000008792 UP000007801 UP000000304 UP000000803 UP000192221 UP000008711 UP000002282 UP000076408 UP000030765 UP000001819 UP000008744 UP000268350 UP000019118 UP000030742 UP000183832 UP000001070 UP000075920 UP000092553 UP000075884 UP000015103 UP000075881
UP000027135 UP000242457 UP000005203 UP000000311 UP000036403 UP000235965 UP000008820 UP000053105 UP000005205 UP000053097 UP000078492 UP000075809 UP000078542 UP000069940 UP000249989 UP000002320 UP000053825 UP000008237 UP000095301 UP000002358 UP000078540 UP000215335 UP000095300 UP000092445 UP000092443 UP000092460 UP000092444 UP000078200 UP000037069 UP000000673 UP000091820 UP000007755 UP000078541 UP000092461 UP000075880 UP000007798 UP000001292 UP000009192 UP000008792 UP000007801 UP000000304 UP000000803 UP000192221 UP000008711 UP000002282 UP000076408 UP000030765 UP000001819 UP000008744 UP000268350 UP000019118 UP000030742 UP000183832 UP000001070 UP000075920 UP000092553 UP000075884 UP000015103 UP000075881
Interpro
SUPFAM
SSF48113
SSF48113
Gene 3D
ProteinModelPortal
H9JRK3
A0A0N0PA47
A0A212EYQ2
A0A0T6B316
A0A1W4W5F5
A0A1Y1KF46
+ More
D6WBT3 A0A154PLF8 A0A067RKW1 A0A2A3EKW9 A0A087ZTY5 E2AB89 A0A0J7KV41 A0A310SWP3 A0A2J7Q6E0 Q16P69 A0A0N0U5W3 A0A158NYM0 A0A026WCF1 A0A195DEI9 A0A151WK86 A0A195CFF8 A0A182GW90 B0WFV6 A0A0L7RI44 A0A0C9PU88 E2B9D0 T1PI01 A0A1Q3EWJ2 A0A0A1WJF6 W8BRE3 A0A034WGB4 K7J6K3 A0A195BS51 A0A034WFA1 A0A232F0H9 A0A1I8NWM7 A0A1B0A6A0 A0A1A9Y2T9 A0A2M4BFL8 A0A1B0AWD4 A0A1B0FEZ3 A0A1A9VKW5 A0A0L0BNG6 W5JX69 A0A2M4APY7 A0A1Y1KMQ2 A0A1I8M8Y6 A0A1A9WSR4 F4W9P9 A0A195FGR7 A0A1B0CKP2 A0A2S2QBA2 A0A0A1XA67 A0A1I8NWN0 A0A224XG85 W8BX98 A0A023F2J9 A0A2H8TLC1 A0A182IZR2 B4ML95 B4IIS2 B4L0W5 B4LBL7 B3M8T4 A0A0Q9WPC4 A0A0Q9XR12 B4QRC8 Q9VW52 A0A1W4W6B9 A0A0Q9WU09 B3NE03 B4PFW5 A0A182YDW1 A0A084VJH2 Q2LYV5 B4H035 A0A0A9W7U3 A0A3B0J5J8 N6UNA6 A0A1L8DGI3 N6TQT2 A0A1J1HE94 A0A0J9RYA9 A0A0P8XW31 Q8MRG6 B4IXN8 A0A1W4VTR7 A0A0Q5UGC8 A0A0R1E0P9 A0A182W7Z7 A0A0M3QWS9 A0A182N8D3 N6UQ17 T1I1M9 A0A182JR22 A0A069DV86 V5IG47 T1J0D7
D6WBT3 A0A154PLF8 A0A067RKW1 A0A2A3EKW9 A0A087ZTY5 E2AB89 A0A0J7KV41 A0A310SWP3 A0A2J7Q6E0 Q16P69 A0A0N0U5W3 A0A158NYM0 A0A026WCF1 A0A195DEI9 A0A151WK86 A0A195CFF8 A0A182GW90 B0WFV6 A0A0L7RI44 A0A0C9PU88 E2B9D0 T1PI01 A0A1Q3EWJ2 A0A0A1WJF6 W8BRE3 A0A034WGB4 K7J6K3 A0A195BS51 A0A034WFA1 A0A232F0H9 A0A1I8NWM7 A0A1B0A6A0 A0A1A9Y2T9 A0A2M4BFL8 A0A1B0AWD4 A0A1B0FEZ3 A0A1A9VKW5 A0A0L0BNG6 W5JX69 A0A2M4APY7 A0A1Y1KMQ2 A0A1I8M8Y6 A0A1A9WSR4 F4W9P9 A0A195FGR7 A0A1B0CKP2 A0A2S2QBA2 A0A0A1XA67 A0A1I8NWN0 A0A224XG85 W8BX98 A0A023F2J9 A0A2H8TLC1 A0A182IZR2 B4ML95 B4IIS2 B4L0W5 B4LBL7 B3M8T4 A0A0Q9WPC4 A0A0Q9XR12 B4QRC8 Q9VW52 A0A1W4W6B9 A0A0Q9WU09 B3NE03 B4PFW5 A0A182YDW1 A0A084VJH2 Q2LYV5 B4H035 A0A0A9W7U3 A0A3B0J5J8 N6UNA6 A0A1L8DGI3 N6TQT2 A0A1J1HE94 A0A0J9RYA9 A0A0P8XW31 Q8MRG6 B4IXN8 A0A1W4VTR7 A0A0Q5UGC8 A0A0R1E0P9 A0A182W7Z7 A0A0M3QWS9 A0A182N8D3 N6UQ17 T1I1M9 A0A182JR22 A0A069DV86 V5IG47 T1J0D7
PDB
6QX9
E-value=2.83468e-121,
Score=1116
Ontologies
GO
PANTHER
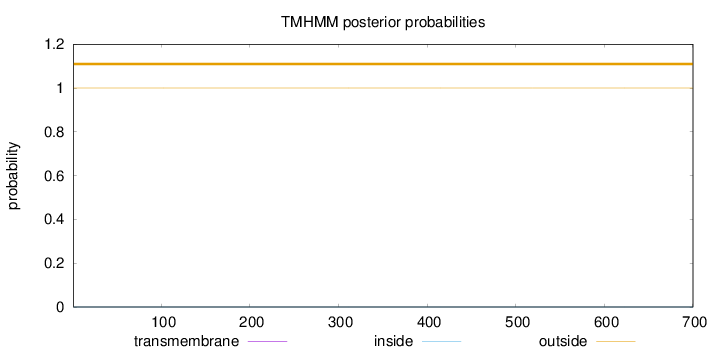
Topology
Length:
700
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00252
Exp number, first 60 AAs:
0.00023
Total prob of N-in:
0.00021
outside
1 - 700
Population Genetic Test Statistics
Pi
156.535847
Theta
140.33833
Tajima's D
0.573336
CLR
0.7373
CSRT
0.540422978851058
Interpretation
Uncertain
