Gene
KWMTBOMO14858 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012159
Annotation
PREDICTED:_protein_hu-li_tai_shao_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.119
Sequence
CDS
ATGGCTGAGACGGACTCAGAGACGATCCCGAACGGCACCGGGCCCCTGTCCACCGAGGAGGAAGACGAGCGCTTGAAGCAGCGTCCGGCCGACATCGACGCCGATGTCCGCGAAATGGAACGCCGGAAGCGCGTCGAAGCTCTCATGTCCTCGAAGCTGTTCCGCGAAGAATTGGAACGGGTCCTCGACCAGCAGATGCACGAGGGCGGCGACGCTCCGCTCCTGCAGAGGATCAAGGAGATGGTCGGCGGGAGACTGCACACCGGAAGCCTCAGGGGTCCTAGCTGCGTTTTGCCGATCAACGACATCCGAGGTATCGAAGGCATAGGGTATGAGAAGGGGGAGAAGATACTCCGCTGCAAGCTGGCCGCCGTGTACCGGCTGGTGGATTTGTTTGGTTGGACGACAACCGGAGTGACGGGGCAGATAACGGCCCGTCTGAACACGGCCGAGGAGCAGGTGCTGACGTCGCCGCGCGGTCTGCTGCCCCACGAGGTGACCGCCTCGTCTCTGGTTAAGGTGGACATGCAGGGCGTCGTCCAGGACCAGGGCACCACCAACTTTCCGGTCAACGTCGAAGGTTTCTCGGTGCACGCGTCCGTGCACGCGGCCCGTCCGGACCTGCGCTGCGTGGTGCACGTGCGCGCGCCCTCCGCTCTCGCCGTGTCCGCCACGCGGCGCGGCGTCCTGCCCCTATGCCAGGAGGCGGCCCTTCTCGGCGAGGTCGCGTACCACACTCTACCCTACGGCGTGCTGGACAACGCCGAGCGCGACAAGCTGGTCCGCGCGCTGGGCCCGCACGCCAAGGTCGTGGTGCTGAGCGGGGCCGGAGCGCTGTGCTGCGGTGCCACGCTGGAGGAGGCCTTCCTGCACGCGCGTCTGCTCACCGCCGCCACCGACGCGCAACTGAAGTTAGCAGCCACGCCGCTCGATGACCTCATCCTGCTTGACGATGAAACGCAGAGACAGATGTACGAGGCGTCCCGCAAGCCGCCCGCCGACGCGAGCAAGTGGCGCGTGGGCGGCGAAGAGTTCGAGGCTCTGATGCGGATGCTGGACAACGCCGGCTTCCGCACCGGCTACATCTACAGGCATCCCCTCATCAAGAACGACGTGCCGAGACCCAAGAATGACGTGGAAGTTCCACCAGCCGTGTCGTCCCTCGGCTATCTGCTTGAAGAGGAGGAGCTGTACAAACAAGGGTTGTGGAAGAAAGGTCAGGGGAAGACCGGTGAGCGAACCCGCTGGCTGAACTCTCCGAACGTCTACCAGAAGGTCGAGGTTCTGGAGACTGGCACCAGCGACCCCAAGAAGATCACTAAGTGGGTGGACGTTAACAACGAAGAGTGGATCCAAGATGGCTCGGCTCAATCGAGTACGCCCGTTAAAATCGATACATTGCAATTCGTACCGAAAAATACGAACCCAAAGGAATTTAAGAAGCTTCAACAACAGATCAAAGAGAATCGCCGCGCGGACAAGATAAGCGCCGGTCCTCAGTCCCACATCCTCGAGGGGGTGACCTGGGACGAAGCCAGCAAGATGGTCGGCGAGGACGCCGTGAACACCCACACGGGGGACCATGTGGTGCTGATGGGAGCGGCCTCGAAGGGCATCATCCAGCGCGGCTACCAGCACAACGCGGCCGTGTACAGCGCGCCCTACGCCCGGAACCCCTTCGACCATATCACCGACAACGAGATCGACGAGTACCGGCGCGACGTCGAGCGCAAGCGGCGCGGGAACGAATACGACACTGACCTGTCCGAATCGGAGGCGATAAGCGCGGCGCAGATGACGTCACCGGCCAGCGCCCCGTCCGAGACAGAAGAGGAGTCGCGCGACGAACACCGAGTTTTGAGAATAGAGACCAAGCAAGCGCCGGTCCGAAGCCAACCAGAAGTGGTCCTGAGCGATGTTGATACAACTGATTTCTTGAACGCCGAGAGAGCCCATGTCGACAGTACTAGAGGTAATCAAGATACTAACACGCTAATTGAAGCTCTCGCCTCTAATAATAGAAACCTCCACGATTATTCCCGTAGCGCTAATTTCGATAACATAATTAGGGTCTATGAAGATTCAGAGAATCGATCGAAACGCAAACGTAATCTTAGCGAAAACTTCCGAAACGCCCACGACAGTAGTGTGTCCACTTTGCGTTCGGTCACTTCATTAGATTTAGACTCGTTAAAATCTAGGTCGTTACCCAGGAGACTTAATTCGAGCGTCTACCGGAGTCGCTCTCACAGTGGTCTGGTCAACTCTTACATTGAGAATCCAATGCCATTTCATTATCACATTCCTAAGTGTTACTCATGCGGTGCGATTAGCGTCCCGTATTCTTGGAACATGAAATTCAACAATAAGAAACTGTCAAAAGACAATTCCGTATACTATTCGACAGAAAACATAAATCAGAGTGTTGAAAAGAAAATAGACATAGTTAGAGTCAAAGAGAATGTTATCCCGATTTACAGAATAGCAAATTCGGAAACAGAATTCGTTGATAGTCAAGAGTTTAGTGAAGATAATTACATTACTGATGTTGGAGAGGGCTTCGCAGTTCACAATAACGTAGTTACCGCCAAAGATATAGATAAGAATGAGAATAAAATGTCGGCTGATATAAGAGATGTGTCTTTGAGTTCAGAAGGCGAATACCACAGTTTTACAGACGACTTTGAGGAAGCCTATGAAAGTCCAGTTAAAAATTTAGTTAATAAAGACATTCGCGACTACGCGGTACCCATTAACTGCTATTTTCCTGATTTCGCAAAGCAAAGTCCCAGGAAATCTCCCTTAAAAACTCAGAATAACATTCTAGAACCTATACTGGAAGAATCGAAGAGTTCGTATGGTGAGGAATCTAATAAATCAACGGATCGTAGTAAGAGTGATTCAGAAATTGATAAGTTCACAGAGATTAGTATAAATGATGTCTTCGATACCGCATTCATAACTAATAATAATAATAATGTTGGTAATAGTAATAATGAAAATAACCCTACGATTGTTGTGAACGTAAACAATACTGTGACAGCTTCTGGAGTTGACAAAGACAATTCAGAACAGAGTAGTAAAATGCCAACAGAAATCACCAGCTTCAATTCGACATACGAGTTTGAAAAATACGAAATGATAACGGAAATTCTCATTTCACTATTCAATTCTATAGATTTTGATTTGATCACAAAGAACAGAGAAAAGTCCATTAGTAGTTTTGATGATAGCAAGGATGGTGTAAGTTTCTCTATAACCGAAATGGTTGAAGAATACGCAATTCAATTTGATAACACTAATATAACTGATTCGTTTGAAGATCACACCAAAGAATCTCAAATAATTGTAGAAGGCGTAATATATTATATATACAATAATGTCTTTTACGAACTCGAACAGAAAAATGGGAAATCTAGACAAACAAAAACCGTCTTCACTGTCGCGGATAGCGAAGATATAATGTACACAGCGATTAATATATTTAATAGTCAGGATATAGATTGTGAATGTGATAACGACAATTCTATAGTTAATGAAGAGAGGGATCAAGATTATAATGAGGATAATTATTCAAATTCAGATATAGCCAATCATAGCTATACAATAGCAGAGGAATTGGTCTCTGAGTTAGTTGATACATCTACAGGCGAATCTAAAGATTTGAATGTAGCTTTTGTTAATACGAGAGATGATCACGATGCTGTATTGAATAGTTTTGGCGGTACATCTAAAATTCTTTCGATGTCACTGACCGAGAATAGTGGAGGTATAAAATATTGGATATCTTTTGACGATTGCGATACAACAGAGGATATTCAATTCGGTTTCAGACGCAGGAGCACCGAGATGCCCAGTTTTATTGTTATCGATAGTAAGATCGAGAAACGCGTGACGAAAAATCATTCGAGTGACAATGAAGGCGAATCGAATGATATTCCTTTCGATTTGAATGCGATTGGGAATGATATCGATGTGCCGAAGAAGAAATGTATTTATGAACTGCGCAAAAGGGAGTATAGTAGCTGGCCTCCGTTTGAAGATACGTTGTTTTATAAGATCATTTCGAACTTCCGAATGTCGGATAGCTTTGACGCCAGCGAATTAGATAGGGTCAGGATCGACAATAGCTTAGGCTGA
Protein
MAETDSETIPNGTGPLSTEEEDERLKQRPADIDADVREMERRKRVEALMSSKLFREELERVLDQQMHEGGDAPLLQRIKEMVGGRLHTGSLRGPSCVLPINDIRGIEGIGYEKGEKILRCKLAAVYRLVDLFGWTTTGVTGQITARLNTAEEQVLTSPRGLLPHEVTASSLVKVDMQGVVQDQGTTNFPVNVEGFSVHASVHAARPDLRCVVHVRAPSALAVSATRRGVLPLCQEAALLGEVAYHTLPYGVLDNAERDKLVRALGPHAKVVVLSGAGALCCGATLEEAFLHARLLTAATDAQLKLAATPLDDLILLDDETQRQMYEASRKPPADASKWRVGGEEFEALMRMLDNAGFRTGYIYRHPLIKNDVPRPKNDVEVPPAVSSLGYLLEEEELYKQGLWKKGQGKTGERTRWLNSPNVYQKVEVLETGTSDPKKITKWVDVNNEEWIQDGSAQSSTPVKIDTLQFVPKNTNPKEFKKLQQQIKENRRADKISAGPQSHILEGVTWDEASKMVGEDAVNTHTGDHVVLMGAASKGIIQRGYQHNAAVYSAPYARNPFDHITDNEIDEYRRDVERKRRGNEYDTDLSESEAISAAQMTSPASAPSETEEESRDEHRVLRIETKQAPVRSQPEVVLSDVDTTDFLNAERAHVDSTRGNQDTNTLIEALASNNRNLHDYSRSANFDNIIRVYEDSENRSKRKRNLSENFRNAHDSSVSTLRSVTSLDLDSLKSRSLPRRLNSSVYRSRSHSGLVNSYIENPMPFHYHIPKCYSCGAISVPYSWNMKFNNKKLSKDNSVYYSTENINQSVEKKIDIVRVKENVIPIYRIANSETEFVDSQEFSEDNYITDVGEGFAVHNNVVTAKDIDKNENKMSADIRDVSLSSEGEYHSFTDDFEEAYESPVKNLVNKDIRDYAVPINCYFPDFAKQSPRKSPLKTQNNILEPILEESKSSYGEESNKSTDRSKSDSEIDKFTEISINDVFDTAFITNNNNNVGNSNNENNPTIVVNVNNTVTASGVDKDNSEQSSKMPTEITSFNSTYEFEKYEMITEILISLFNSIDFDLITKNREKSISSFDDSKDGVSFSITEMVEEYAIQFDNTNITDSFEDHTKESQIIVEGVIYYIYNNVFYELEQKNGKSRQTKTVFTVADSEDIMYTAINIFNSQDIDCECDNDNSIVNEERDQDYNEDNYSNSDIANHSYTIAEELVSELVDTSTGESKDLNVAFVNTRDDHDAVLNSFGGTSKILSMSLTENSGGIKYWISFDDCDTTEDIQFGFRRRSTEMPSFIVIDSKIEKRVTKNHSSDNEGESNDIPFDLNAIGNDIDVPKKKCIYELRKREYSSWPPFEDTLFYKIISNFRMSDSFDASELDRVRIDNSLG
Summary
Uniprot
Pubmed
Proteomes
PRIDE
ProteinModelPortal
PDB
3OCR
E-value=3.40068e-22,
Score=265
Ontologies
GO
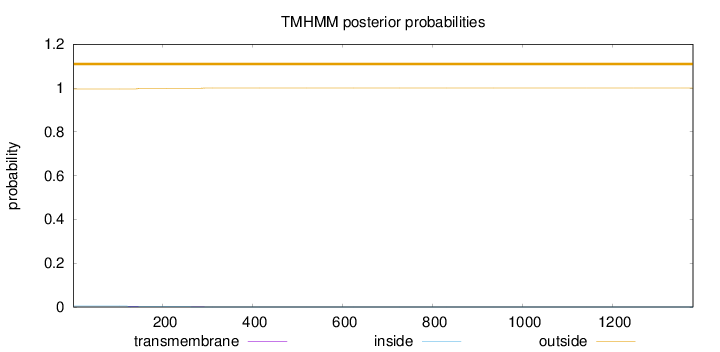
Topology
Length:
1379
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0902999999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00402
outside
1 - 1379
Population Genetic Test Statistics
Pi
240.31193
Theta
156.660892
Tajima's D
2.301116
CLR
0.194475
CSRT
0.92120393980301
Interpretation
Uncertain
