Gene
KWMTBOMO14855
Pre Gene Modal
BGIBMGA012158
Annotation
PREDICTED:_AT-rich_interactive_domain-containing_protein_5B-like_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 3.599
Sequence
CDS
ATGAAACGGGCCCGAACGACGAAGCACACGAATATTGAGGAAGCTTTGCTGAAGTGGTTCAAATATCAGAGGTCTAATAATGTGCCAATAAACGGACCGATTCTACAAGAAAAAGCTAACCATTTTGCTAAATGTTTTGGCGAAGATTTTGTTTGCTCGTCTAGTTGGATCCAACGTTTTCGGGCTCGGCACGGTATTGTCGGCGGGAAAATGAGTCGTGAAGCTGCTAGTGTTGATAAGGATACTGTAGAAGAATGGAAAACGCAAAAGCGGCCTACATTATTTGAAGGCTACGGACCAGATGAAGTTTTTAACGCAGCCGAGACTGGACTTTTTTACAATATGACACCAGACAAGACGTTAAAGTTCAAAGGAGAGAATTGTTCGGGAGGAAAAATGTCTAAGACAAGATTAACGATCATGGTCGCAGCAAATATGACAGGATCATGTAAAAGGAAGCTTTTAGTTATTGGCGAATCTAAGAAACCAAAATGCTTTAAAAACACACACTCACTACCGGTAACGTATGAAAACAATGTGCAGTCATGGATGACATCGGACATTTTTGAGAGATGGTTAAGAAACTGGGACGCTGAATTGAAAGGAAACAAACAGAAAGTTCTACTTTTGGTTGATAACTGTCCTGCTCATCCGGCTGTTACTAATCTAAAATGCATTAAACTTGTGTTCTTACCGCCAAATGTTACATCTGTATTGCAACCAATGGAGCAAGGTGTAATAAGGTGTTTAAAATCTCACTATAGAAGACTGCAAGTGTTAAAGTTAATACAAAACCTTTACAGTGGCGACCAAAAAAGCTTTACGATTCTTGATGCCATCTTGATGATATCAGAAGCATGGAAAAATGTTTCACAAAACACTATAGCTAACTGCTTTAAACATGCTGGTTTTAAAGACCTCTTAACAACAGCCTCAGATGATGTGACTGATGATGATGGAGAGGACAATATTTCTCTTGCTCAATTATCCCAAAATTTACGACCTGCTTTATCTACTACTGAAGCAGCAGAGTTTATTGATGTAGATAACTCTATTGCAATTTGCGCACGAGCTACGGAAGATGATATTGTACGAGATGTTCAAGAGGAGGATGACGAACAGGAAGATTCTGAAGAAAAATTTACTGTACCAACTTTGAATGATGGTATCAATGCGGACACGTTCGAATACCCGGAGCTCGAGGGTCACGAGTTCGTCTGCAACCACCTCGCTCCGCGGCTGAAAGGCAGGCCGAGGGGACGGCGACGGCGCGCAGCCAGGTCCAGGTCGCACTCGCCCTCGCCCTCCTCCCGGTCCAGCGACTCGGACCAGCAGAGCGGCAACCCTGAGCCCGATACGCCGCGGCGCCTCTCTCTGCGGAACGGCGCCGATCGGAACAGCGACGACGAGAACGAGTACCGGCGGCAGGAGACGCCCGAGGACCGGGCCTTCGTCAACCAGCTCAGGCAGTTCTACAGGGACAGGAACGAGACCTTCAAGGAGCACGCCCTCAAAGATGTGACGCCTTTGGCCCTGTACCGCGGGGTGTGCTCGCGGGGCGGCTACCGGGCCGTGTGCCGCCGCGAGCTGTGGCCGCGCCTCCTGCCGCGCCCGGCGCTCGCCCGACGGTACTATGAGAGGTACCTCCTCCCCTTCGAGACCTACGAGCGGAAACTGAACGGCAGCACCGAAAACATCCCGACCATCGAGGTAGCCGACTCCCCACAGCACTCGCCCAGCAAGAAGCCGCTGGACAGGACGAGGGTCATCGATGTGGACGACGAAACGGGCGAGGTCAAGGACGTGCCCGACTGCCCCGCCACCAGCCAGCCCGCCGAGGTGCTCAACCGGGAGTTCCTGGACTCGCTGGACGCGGACAGGCGCGCCGCCAAGATCAACGTCGTGCCCGTCGAGAAGCTGAAGGACGAGAAATGCGCCGTCATCAACAACAACGTGCACCTGCCGAACACATCATCGAACGCTGAACAGACGATAGTGAGGACGAACCCTGTGACCACCGAGCCGGTTGTCAACGGGCATGCGGACAATGGACCGCAGAAGACGGTCCGTCCCCTCGGTAAGAGTTCCCTCCGCGCCGTCCGAGTGAAGGCGACGCGACCTCACGCGCAGCCCGCCAGTGCACCTCCGTCGCTCCTGAGGGGATCCCCGCCCGCCGCCGGACTCAACAACTTCGGCATCCACCGGCCCCCGCCCGCCCCGCGCCGCGCCCCTCGCAGCGACGACGAGATCGTGGAGGTCCCGTACAAGCCCAAGAGTCCCGAGATCATAGACCTGGACGAGTACCCGGAGAGTCCTCAGACCTTCAAGAAGAAGAAGCTGGAGATACTCAAGGAGCGAGGACTGGAGGTGACGGTGCTGCCCGCGGGCCCCTGGCCGGTGCTGGCGCCCGTCGCGGCCGTGCAGCACCTCGGCTACCCGCGGCCCGCGCAGCTGTACCAGGTGTTCGGCGCGGGCCCGCCCAAGGTCATCCAGTCCACCTCGCCCTTCGGCTGCGACGGGCCCCAGAAGACGGTCTACGGGAACCCTAAGGACCCGTTCATGCCGCCGCCGCACGTGCTGCAGGGCGCGCCCGTGAGGCCGCAGCGAGCCCCCCCCGCCCCCCACGAGCCGCCGCACGTGCTCGACCTCACGTGCAAGCTGGGGTCGCCCGCGGGCAGCGCCGTCCGCGTCGACTCTGATCTGGAGATCACGCTGCTTGAGGACACGAACGCCTCGCAGAAGAGGCCGGCGCTCCGCCCCCCGTGCGGCGCGGACGGGGCGGTCGGCTACAGCTACAAGGACCCGTACGCCGCCTTCTACAACAGCCTGGCGGGGTCCGTGGACCAAAGGCAGCTGGCCATGTACCGAGAGCTGGTCTCCACGCAGTTCCGCGGCTACGGGCTGGCGGGCCTCGCGCCCGGACACGTGCCCACCAGCAAGTGA
Protein
MKRARTTKHTNIEEALLKWFKYQRSNNVPINGPILQEKANHFAKCFGEDFVCSSSWIQRFRARHGIVGGKMSREAASVDKDTVEEWKTQKRPTLFEGYGPDEVFNAAETGLFYNMTPDKTLKFKGENCSGGKMSKTRLTIMVAANMTGSCKRKLLVIGESKKPKCFKNTHSLPVTYENNVQSWMTSDIFERWLRNWDAELKGNKQKVLLLVDNCPAHPAVTNLKCIKLVFLPPNVTSVLQPMEQGVIRCLKSHYRRLQVLKLIQNLYSGDQKSFTILDAILMISEAWKNVSQNTIANCFKHAGFKDLLTTASDDVTDDDGEDNISLAQLSQNLRPALSTTEAAEFIDVDNSIAICARATEDDIVRDVQEEDDEQEDSEEKFTVPTLNDGINADTFEYPELEGHEFVCNHLAPRLKGRPRGRRRRAARSRSHSPSPSSRSSDSDQQSGNPEPDTPRRLSLRNGADRNSDDENEYRRQETPEDRAFVNQLRQFYRDRNETFKEHALKDVTPLALYRGVCSRGGYRAVCRRELWPRLLPRPALARRYYERYLLPFETYERKLNGSTENIPTIEVADSPQHSPSKKPLDRTRVIDVDDETGEVKDVPDCPATSQPAEVLNREFLDSLDADRRAAKINVVPVEKLKDEKCAVINNNVHLPNTSSNAEQTIVRTNPVTTEPVVNGHADNGPQKTVRPLGKSSLRAVRVKATRPHAQPASAPPSLLRGSPPAAGLNNFGIHRPPPAPRRAPRSDDEIVEVPYKPKSPEIIDLDEYPESPQTFKKKKLEILKERGLEVTVLPAGPWPVLAPVAAVQHLGYPRPAQLYQVFGAGPPKVIQSTSPFGCDGPQKTVYGNPKDPFMPPPHVLQGAPVRPQRAPPAPHEPPHVLDLTCKLGSPAGSAVRVDSDLEITLLEDTNASQKRPALRPPCGADGAVGYSYKDPYAAFYNSLAGSVDQRQLAMYRELVSTQFRGYGLAGLAPGHVPTSK
Summary
Uniprot
Proteomes
PRIDE
Interpro
Gene 3D
ProteinModelPortal
PDB
1HLV
E-value=0.000133614,
Score=112
Ontologies
GO
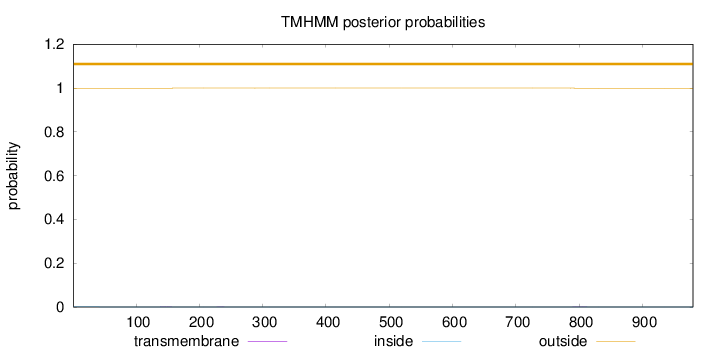
Topology
Subcellular location
Nucleus
Length:
980
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01752
Exp number, first 60 AAs:
0.00052
Total prob of N-in:
0.00059
outside
1 - 980
Population Genetic Test Statistics
Pi
243.521997
Theta
195.86455
Tajima's D
1.287024
CLR
0.409414
CSRT
0.739713014349282
Interpretation
Uncertain
