Pre Gene Modal
BGIBMGA012193
Annotation
PREDICTED:_glycerol-3-phosphate_acyltransferase_4_isoform_X3_[Papilio_xuthus]
Full name
Glycerol-3-phosphate acyltransferase 3
Alternative Name
1-acyl-sn-glycerol-3-phosphate O-acyltransferase 9
Acyl-CoA:glycerol-3-phosphate acyltransferase 3
Lysophosphatidic acid acyltransferase theta
Acyl-CoA:glycerol-3-phosphate acyltransferase 3
Lysophosphatidic acid acyltransferase theta
Location in the cell
Mitochondrial Reliability : 1.442 PlasmaMembrane Reliability : 1.555
Sequence
CDS
ATGATAATCACAACGACGCTGGTCAGCACGCTACCGTCTGGTAGGGTGAGGCGTTACCTCGGCGCTGTCTCCTACAAAATCTCCATCCGCGTACTGATAAGGAGTCTGTCGTGCGTGATAACGTTCCACGACCAGCAGCACATGCCGAAATCGAACGGCTTCTGTGTCGCCAATCACACGAGCCCGATCGATGTGGCCATCGTCAGCGTGAACACGTGCTTCTCATTGATTGGACAGAGGCACGGAGGATTCCTCGGTCTCCTACAGAGGGCTCTCGCGAGGGCGTCCCCTCACATTTGGTTCGAGAGGTCGGAGGTTAAAGACAGGCACGCCGTCGCGAGGAGACTAAAAGAACACATATCGGTTGCCGACAATCCTCCTATACTGATATTTCCCGAAGGCACCTGCATAAATAACACGTCGGTGATGCAGTTCAAGAAGGGCAGCTTCGAAGTCGGCGGCACTGTCTACCCAGTGGCAATCAAGTGA
Protein
MIITTTLVSTLPSGRVRRYLGAVSYKISIRVLIRSLSCVITFHDQQHMPKSNGFCVANHTSPIDVAIVSVNTCFSLIGQRHGGFLGLLQRALARASPHIWFERSEVKDRHAVARRLKEHISVADNPPILIFPEGTCINNTSVMQFKKGSFEVGGTVYPVAIK
Summary
Description
May transfer the acyl-group from acyl-coA to the sn-1 position of glycerol-3-phosphate, an essential step in glycerolipid biosynthesis. Also transfers the acyl-group from acyl-coA to the sn-2 position of 1-acyl-sn-glycerol-3-phosphate (lysophosphatidic acid, or LPA), forming 1,2-diacyl-sn-glycerol-3-phosphate (phosphatidic acid, or PA).
Catalytic Activity
an acyl-CoA + sn-glycerol 3-phosphate = a 1-acyl-sn-glycero-3-phosphate + CoA
a 1-acyl-sn-glycero-3-phosphate + an acyl-CoA = a 1,2-diacyl-sn-glycero-3-phosphate + CoA
a 1-acyl-sn-glycero-3-phosphate + an acyl-CoA = a 1,2-diacyl-sn-glycero-3-phosphate + CoA
Similarity
Belongs to the 1-acyl-sn-glycerol-3-phosphate acyltransferase family.
Keywords
Acyltransferase
Complete proteome
Endoplasmic reticulum
Lipid biosynthesis
Lipid metabolism
Membrane
Phospholipid biosynthesis
Phospholipid metabolism
Phosphoprotein
Reference proteome
Transferase
Transmembrane
Transmembrane helix
Feature
chain Glycerol-3-phosphate acyltransferase 3
Uniprot
A0A2A4J561
A0A1E1W253
W8C971
A0A0A1X8M2
A0A0K8V513
A0A2A4J4L5
+ More
A0A0L7LGZ1 A0A0K8UU81 A0A1L8EDX2 A0A1L8EDT9 V5GTI7 A0A291P138 A0A291P129 V5GHQ3 A0A2W1BJQ5 A0A1I8M243 A0A2J7PWA8 A0A1I8Q8N5 A0A0R3NTV1 A0A3B0J2H0 A0A1Y1LA95 A0A0N1IAB6 A0A0P0EA76 A0A1V0M890 A0A088M9A3 A0A194Q131 A0A0Q9X8X4 Q9W1B5 B4PAT5 A0A0J9RK61 B3NQ72 A0A1W4UMC0 B3MDX1 A0A1W4XCD5 A0A0C9QPY2 B4LPD3 A0A3S2N4Q7 A0A1L8DY55 W5J2T7 A0A212F2Q5 A0A1B6ECR7 A0A1L8DYS0 A0A2P8YTM3 E0VSI7 A0A1B6HSN4 A0A1Y1LAC1 A0A1Y1LG56 A0A0C9QCN7 A0A1W7R898 A0A224XPG7 A0A154PMK4 A0A1B0FK30 A0A1B0C1H5 A0A067QT47 A0A2J7PWA6 A0A1B6GNX9 A0A1A9VKM6 A0A0L7QVJ6 A0A023ET90 A0A232FLL4 A0A1A9XAC6 K7IT41 A0A1A9ZVJ6 A0A0V0G3F7 D6WBN5 T1IW46 A0A0U3BIT0 A0A060YX47 A0A0M9ACA5 T1HC21 A0A1S4EVW1 G1KKZ9 A0A2D4KFS3 A0A0T6AZD4 U3FCN4 E2A055 T1P9H1 A0A2A3EJP6 A0A088AGA7 A0A182GZ08 A0A1Q3FVU1 A0A250YI92 A0A1Q3FVT2 B0W6P5 A0A151P208 A0A0B8RXL7 A0A1U7SBP5 A0A3L8DI46 A0A0J7L5L0 G3IK30 A0A1Q3FW83 A0A1Y1LF29 A0A1Q3FVT1 A0A1Q3FVU0 A0A1Q3FWB7 Q17P93 A0A146KKN9 A0A1S3FHJ5 K7GAT1 Q8C0N2
A0A0L7LGZ1 A0A0K8UU81 A0A1L8EDX2 A0A1L8EDT9 V5GTI7 A0A291P138 A0A291P129 V5GHQ3 A0A2W1BJQ5 A0A1I8M243 A0A2J7PWA8 A0A1I8Q8N5 A0A0R3NTV1 A0A3B0J2H0 A0A1Y1LA95 A0A0N1IAB6 A0A0P0EA76 A0A1V0M890 A0A088M9A3 A0A194Q131 A0A0Q9X8X4 Q9W1B5 B4PAT5 A0A0J9RK61 B3NQ72 A0A1W4UMC0 B3MDX1 A0A1W4XCD5 A0A0C9QPY2 B4LPD3 A0A3S2N4Q7 A0A1L8DY55 W5J2T7 A0A212F2Q5 A0A1B6ECR7 A0A1L8DYS0 A0A2P8YTM3 E0VSI7 A0A1B6HSN4 A0A1Y1LAC1 A0A1Y1LG56 A0A0C9QCN7 A0A1W7R898 A0A224XPG7 A0A154PMK4 A0A1B0FK30 A0A1B0C1H5 A0A067QT47 A0A2J7PWA6 A0A1B6GNX9 A0A1A9VKM6 A0A0L7QVJ6 A0A023ET90 A0A232FLL4 A0A1A9XAC6 K7IT41 A0A1A9ZVJ6 A0A0V0G3F7 D6WBN5 T1IW46 A0A0U3BIT0 A0A060YX47 A0A0M9ACA5 T1HC21 A0A1S4EVW1 G1KKZ9 A0A2D4KFS3 A0A0T6AZD4 U3FCN4 E2A055 T1P9H1 A0A2A3EJP6 A0A088AGA7 A0A182GZ08 A0A1Q3FVU1 A0A250YI92 A0A1Q3FVT2 B0W6P5 A0A151P208 A0A0B8RXL7 A0A1U7SBP5 A0A3L8DI46 A0A0J7L5L0 G3IK30 A0A1Q3FW83 A0A1Y1LF29 A0A1Q3FVT1 A0A1Q3FVU0 A0A1Q3FWB7 Q17P93 A0A146KKN9 A0A1S3FHJ5 K7GAT1 Q8C0N2
EC Number
2.3.1.15
Pubmed
24495485
25830018
26227816
28986331
28756777
25315136
+ More
15632085 17994087 28004739 26354079 26445454 28349297 26385554 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22936249 20920257 23761445 22118469 29403074 20566863 24845553 24945155 28648823 20075255 18362917 19820115 24755649 23915248 20798317 26483478 28087693 22293439 25476704 30249741 21804562 17510324 26823975 17381049 16141072 15489334 17170135 17242355 18630941 19144319 21183079
15632085 17994087 28004739 26354079 26445454 28349297 26385554 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22936249 20920257 23761445 22118469 29403074 20566863 24845553 24945155 28648823 20075255 18362917 19820115 24755649 23915248 20798317 26483478 28087693 22293439 25476704 30249741 21804562 17510324 26823975 17381049 16141072 15489334 17170135 17242355 18630941 19144319 21183079
EMBL
NWSH01003180
PCG66818.1
GDQN01010040
JAT81014.1
GAMC01006896
JAB99659.1
+ More
GBXI01006638 JAD07654.1 GDHF01018292 JAI34022.1 PCG66819.1 JTDY01001227 KOB74476.1 GDHF01022226 JAI30088.1 GFDG01001933 JAV16866.1 GFDG01001932 JAV16867.1 GALX01004913 JAB63553.1 MF706175 ATJ44602.1 MF687643 ATJ44569.1 GALX01004912 JAB63554.1 KZ150102 PZC73497.1 NEVH01020936 PNF20613.1 CM000071 KRT02243.1 OUUW01000001 SPP73233.1 GEZM01061280 JAV70572.1 KQ460396 KPJ15331.1 KT261717 ALJ30257.1 KU755496 ARD71206.1 KJ579236 AIN34712.1 KQ459580 KPI99261.1 CH933808 KRG04891.1 AE013599 BT072873 AAF47157.2 ACN63495.1 CM000158 EDW92475.2 CM002911 KMY96251.1 CH954179 EDV56945.2 CH902619 EDV37516.2 GBYB01005709 JAG75476.1 CH940648 EDW61192.2 RSAL01000568 RVE41378.1 GFDF01002738 JAV11346.1 ADMH02002129 ETN58607.1 AGBW02010693 OWR48025.1 GEDC01001611 JAS35687.1 GFDF01002642 JAV11442.1 PYGN01000366 PSN47586.1 DS235751 EEB16343.1 GECU01030036 JAS77670.1 GEZM01061279 JAV70574.1 GEZM01061277 JAV70576.1 GBYB01012183 JAG81950.1 GEHC01000271 JAV47374.1 GFTR01006373 JAW10053.1 KQ434980 KZC13082.1 CCAG010017210 JXJN01024053 KK852969 KDR13114.1 PNF20614.1 GECZ01005635 JAS64134.1 KQ414726 KOC62640.1 GAPW01001058 JAC12540.1 NNAY01000061 OXU31399.1 AAZX01006711 GECL01003497 JAP02627.1 KQ971311 EEZ98897.2 JH431607 KT328600 ALT07197.1 FR925089 CDQ96306.1 KQ435706 KOX80219.1 ACPB03008412 ACPB03008413 ACPB03008414 IACL01056014 LAB07571.1 LJIG01022464 KRT80440.1 GAEP01000617 JAB54204.1 GL435551 EFN73188.1 KA644810 AFP59439.1 KZ288235 PBC31406.1 JXUM01021838 JXUM01021839 JXUM01021840 JXUM01021841 JXUM01021842 JXUM01021843 KQ560612 KXJ81628.1 GFDL01003326 JAV31719.1 GFFW01001522 JAV43266.1 GFDL01003324 JAV31721.1 DS231849 EDS36876.1 AKHW03001358 KYO42735.1 GBSH01001173 JAG67852.1 QOIP01000007 RLU19996.1 LBMM01000472 KMQ98257.1 JH003478 EGW11247.1 GFDL01003319 JAV31726.1 GEZM01061278 JAV70575.1 GFDL01003331 JAV31714.1 GFDL01003321 JAV31724.1 GFDL01003332 JAV31713.1 CH477193 EAT48539.1 GDHC01021940 JAP96688.1 AGCU01005067 AGCU01005068 AGCU01005069 AGCU01005070 AGCU01005071 AK030171 AK160261 AK138410 BC096769 BC138228 BC145669
GBXI01006638 JAD07654.1 GDHF01018292 JAI34022.1 PCG66819.1 JTDY01001227 KOB74476.1 GDHF01022226 JAI30088.1 GFDG01001933 JAV16866.1 GFDG01001932 JAV16867.1 GALX01004913 JAB63553.1 MF706175 ATJ44602.1 MF687643 ATJ44569.1 GALX01004912 JAB63554.1 KZ150102 PZC73497.1 NEVH01020936 PNF20613.1 CM000071 KRT02243.1 OUUW01000001 SPP73233.1 GEZM01061280 JAV70572.1 KQ460396 KPJ15331.1 KT261717 ALJ30257.1 KU755496 ARD71206.1 KJ579236 AIN34712.1 KQ459580 KPI99261.1 CH933808 KRG04891.1 AE013599 BT072873 AAF47157.2 ACN63495.1 CM000158 EDW92475.2 CM002911 KMY96251.1 CH954179 EDV56945.2 CH902619 EDV37516.2 GBYB01005709 JAG75476.1 CH940648 EDW61192.2 RSAL01000568 RVE41378.1 GFDF01002738 JAV11346.1 ADMH02002129 ETN58607.1 AGBW02010693 OWR48025.1 GEDC01001611 JAS35687.1 GFDF01002642 JAV11442.1 PYGN01000366 PSN47586.1 DS235751 EEB16343.1 GECU01030036 JAS77670.1 GEZM01061279 JAV70574.1 GEZM01061277 JAV70576.1 GBYB01012183 JAG81950.1 GEHC01000271 JAV47374.1 GFTR01006373 JAW10053.1 KQ434980 KZC13082.1 CCAG010017210 JXJN01024053 KK852969 KDR13114.1 PNF20614.1 GECZ01005635 JAS64134.1 KQ414726 KOC62640.1 GAPW01001058 JAC12540.1 NNAY01000061 OXU31399.1 AAZX01006711 GECL01003497 JAP02627.1 KQ971311 EEZ98897.2 JH431607 KT328600 ALT07197.1 FR925089 CDQ96306.1 KQ435706 KOX80219.1 ACPB03008412 ACPB03008413 ACPB03008414 IACL01056014 LAB07571.1 LJIG01022464 KRT80440.1 GAEP01000617 JAB54204.1 GL435551 EFN73188.1 KA644810 AFP59439.1 KZ288235 PBC31406.1 JXUM01021838 JXUM01021839 JXUM01021840 JXUM01021841 JXUM01021842 JXUM01021843 KQ560612 KXJ81628.1 GFDL01003326 JAV31719.1 GFFW01001522 JAV43266.1 GFDL01003324 JAV31721.1 DS231849 EDS36876.1 AKHW03001358 KYO42735.1 GBSH01001173 JAG67852.1 QOIP01000007 RLU19996.1 LBMM01000472 KMQ98257.1 JH003478 EGW11247.1 GFDL01003319 JAV31726.1 GEZM01061278 JAV70575.1 GFDL01003331 JAV31714.1 GFDL01003321 JAV31724.1 GFDL01003332 JAV31713.1 CH477193 EAT48539.1 GDHC01021940 JAP96688.1 AGCU01005067 AGCU01005068 AGCU01005069 AGCU01005070 AGCU01005071 AK030171 AK160261 AK138410 BC096769 BC138228 BC145669
Proteomes
UP000218220
UP000037510
UP000095301
UP000235965
UP000095300
UP000001819
+ More
UP000268350 UP000053240 UP000053268 UP000009192 UP000000803 UP000002282 UP000008711 UP000192221 UP000007801 UP000192223 UP000008792 UP000283053 UP000000673 UP000007151 UP000245037 UP000009046 UP000076502 UP000092444 UP000092460 UP000027135 UP000078200 UP000053825 UP000215335 UP000092443 UP000002358 UP000092445 UP000007266 UP000193380 UP000053105 UP000015103 UP000001646 UP000000311 UP000242457 UP000005203 UP000069940 UP000249989 UP000002320 UP000050525 UP000189705 UP000279307 UP000036403 UP000001075 UP000008820 UP000081671 UP000007267 UP000000589
UP000268350 UP000053240 UP000053268 UP000009192 UP000000803 UP000002282 UP000008711 UP000192221 UP000007801 UP000192223 UP000008792 UP000283053 UP000000673 UP000007151 UP000245037 UP000009046 UP000076502 UP000092444 UP000092460 UP000027135 UP000078200 UP000053825 UP000215335 UP000092443 UP000002358 UP000092445 UP000007266 UP000193380 UP000053105 UP000015103 UP000001646 UP000000311 UP000242457 UP000005203 UP000069940 UP000249989 UP000002320 UP000050525 UP000189705 UP000279307 UP000036403 UP000001075 UP000008820 UP000081671 UP000007267 UP000000589
Pfam
PF01553 Acyltransferase
Interpro
IPR002123
Plipid/glycerol_acylTrfase
ProteinModelPortal
A0A2A4J561
A0A1E1W253
W8C971
A0A0A1X8M2
A0A0K8V513
A0A2A4J4L5
+ More
A0A0L7LGZ1 A0A0K8UU81 A0A1L8EDX2 A0A1L8EDT9 V5GTI7 A0A291P138 A0A291P129 V5GHQ3 A0A2W1BJQ5 A0A1I8M243 A0A2J7PWA8 A0A1I8Q8N5 A0A0R3NTV1 A0A3B0J2H0 A0A1Y1LA95 A0A0N1IAB6 A0A0P0EA76 A0A1V0M890 A0A088M9A3 A0A194Q131 A0A0Q9X8X4 Q9W1B5 B4PAT5 A0A0J9RK61 B3NQ72 A0A1W4UMC0 B3MDX1 A0A1W4XCD5 A0A0C9QPY2 B4LPD3 A0A3S2N4Q7 A0A1L8DY55 W5J2T7 A0A212F2Q5 A0A1B6ECR7 A0A1L8DYS0 A0A2P8YTM3 E0VSI7 A0A1B6HSN4 A0A1Y1LAC1 A0A1Y1LG56 A0A0C9QCN7 A0A1W7R898 A0A224XPG7 A0A154PMK4 A0A1B0FK30 A0A1B0C1H5 A0A067QT47 A0A2J7PWA6 A0A1B6GNX9 A0A1A9VKM6 A0A0L7QVJ6 A0A023ET90 A0A232FLL4 A0A1A9XAC6 K7IT41 A0A1A9ZVJ6 A0A0V0G3F7 D6WBN5 T1IW46 A0A0U3BIT0 A0A060YX47 A0A0M9ACA5 T1HC21 A0A1S4EVW1 G1KKZ9 A0A2D4KFS3 A0A0T6AZD4 U3FCN4 E2A055 T1P9H1 A0A2A3EJP6 A0A088AGA7 A0A182GZ08 A0A1Q3FVU1 A0A250YI92 A0A1Q3FVT2 B0W6P5 A0A151P208 A0A0B8RXL7 A0A1U7SBP5 A0A3L8DI46 A0A0J7L5L0 G3IK30 A0A1Q3FW83 A0A1Y1LF29 A0A1Q3FVT1 A0A1Q3FVU0 A0A1Q3FWB7 Q17P93 A0A146KKN9 A0A1S3FHJ5 K7GAT1 Q8C0N2
A0A0L7LGZ1 A0A0K8UU81 A0A1L8EDX2 A0A1L8EDT9 V5GTI7 A0A291P138 A0A291P129 V5GHQ3 A0A2W1BJQ5 A0A1I8M243 A0A2J7PWA8 A0A1I8Q8N5 A0A0R3NTV1 A0A3B0J2H0 A0A1Y1LA95 A0A0N1IAB6 A0A0P0EA76 A0A1V0M890 A0A088M9A3 A0A194Q131 A0A0Q9X8X4 Q9W1B5 B4PAT5 A0A0J9RK61 B3NQ72 A0A1W4UMC0 B3MDX1 A0A1W4XCD5 A0A0C9QPY2 B4LPD3 A0A3S2N4Q7 A0A1L8DY55 W5J2T7 A0A212F2Q5 A0A1B6ECR7 A0A1L8DYS0 A0A2P8YTM3 E0VSI7 A0A1B6HSN4 A0A1Y1LAC1 A0A1Y1LG56 A0A0C9QCN7 A0A1W7R898 A0A224XPG7 A0A154PMK4 A0A1B0FK30 A0A1B0C1H5 A0A067QT47 A0A2J7PWA6 A0A1B6GNX9 A0A1A9VKM6 A0A0L7QVJ6 A0A023ET90 A0A232FLL4 A0A1A9XAC6 K7IT41 A0A1A9ZVJ6 A0A0V0G3F7 D6WBN5 T1IW46 A0A0U3BIT0 A0A060YX47 A0A0M9ACA5 T1HC21 A0A1S4EVW1 G1KKZ9 A0A2D4KFS3 A0A0T6AZD4 U3FCN4 E2A055 T1P9H1 A0A2A3EJP6 A0A088AGA7 A0A182GZ08 A0A1Q3FVU1 A0A250YI92 A0A1Q3FVT2 B0W6P5 A0A151P208 A0A0B8RXL7 A0A1U7SBP5 A0A3L8DI46 A0A0J7L5L0 G3IK30 A0A1Q3FW83 A0A1Y1LF29 A0A1Q3FVT1 A0A1Q3FVU0 A0A1Q3FWB7 Q17P93 A0A146KKN9 A0A1S3FHJ5 K7GAT1 Q8C0N2
PDB
5KYM
E-value=0.042898,
Score=81
Ontologies
GO
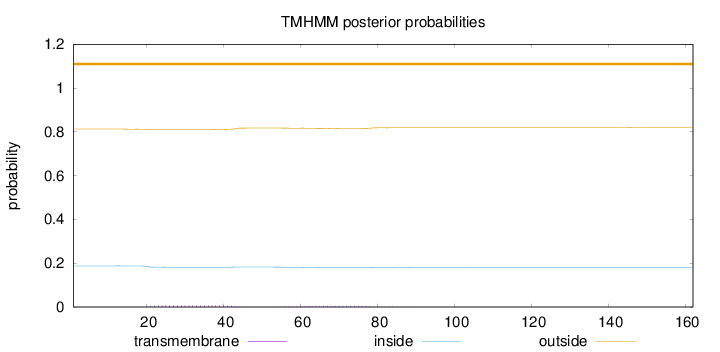
Topology
Subcellular location
Endoplasmic reticulum membrane
Length:
162
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.28611
Exp number, first 60 AAs:
0.18551
Total prob of N-in:
0.18761
outside
1 - 162
Population Genetic Test Statistics
Pi
295.179009
Theta
211.616803
Tajima's D
0.72689
CLR
120.617031
CSRT
0.584220788960552
Interpretation
Uncertain
