Pre Gene Modal
BGIBMGA012193
Annotation
PREDICTED:_glycerol-3-phosphate_acyltransferase_3_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.206 Nuclear Reliability : 1.398
Sequence
CDS
ATGATGAGCTCGTGGGCGATAGTCTGCGACGTGTGGTACCTGCCGGCGATGTCCCGGCGCCCGGACGAGACCGCGGTCGACTTCGCGAACCGGGTCAAGTCGGTGATCGCCAGGCAGGGCGGCCTCATCGACCTGCAATGGGACGGACAGTTAAAGAGAACGAAAGCGAAGAAGGAGTGGAGGGAGCTCCAGCAAGAGGAGTTCAGTAAGAGGTTAAAAGTGGAATAG
Protein
MMSSWAIVCDVWYLPAMSRRPDETAVDFANRVKSVIARQGGLIDLQWDGQLKRTKAKKEWRELQQEEFSKRLKVE
Summary
Uniprot
H9JRN3
K7IT41
A0A195D8P5
A0A195D0G0
A0A158ND16
A0A195B8Y9
+ More
F4X0K5 A0A195F7N3 A0A0C9QPY2 A0A3S2N4Q7 A0A0C9QCN7 A0A1E1W253 E2B6U5 A0A194Q131 A0A1B6HSN4 A0A1B6GNX9 A0A0N1IAB6 A0A2A3EJP6 A0A088AGA7 A0A212F2Q5 A0A026WGX4 A0A154PMK4 A0A0M9ACA5 D6WBN5 A0A0P5A9Q3 A0A0P5DP87 U5EQE7 A0A2P8YTM3 A0A1Y1LAC1 A0A1Y1LF29 A0A1Y1LA95 A0A1Y1LG56 A0A0N8EMW7 A0A1L8DYB1 Q17P93 A0A1L8DYS0 A0A0P6ADJ6 A0A1B0CEV6 B0W6P5 A0A023ET90 A0A2M4BK03 A0A2M4BK52 A0A2M4BK12 A0A2M4BJY5 A0A182J1V5 A0A336K4K1 A0A1B6ECR7 V5GTI7 B4QBI8 A0A232FLL4 A0A0B4LGJ9 W8C971 A0A1I8M244 A0A1E1VYY8 B4I2C7 B4GCJ6 A0A182FRW8 W8BEE7 A0A1I8Q8R5 A0A1I8M243 A0A2H1VUV3 A0A1L8EDT9 A0A0L0CDG6 E0VSI7 A0A1L8EDX2 A0A1I8Q8N5 W5J2T7 A0A3B0JKR7 T1P9H1 A0A1A9XAC6 A0A2M4CZG5 A0A1L8EIS8 A0A1A9ZVJ6 A0A2M4ACX9 A0A1A9VKM6 A0A1B0FK30 A0A2M3Z840 A0A1I8Q8N3 A0A2M3Z7V7 A0A182TYJ3 A0A1W4V042 A0A182QDW4 A0A1B0C1H5 A0A2S2QGS3 A0A1B6JYC1 A0A182XW00 A0A182LR17 B4J6E5 A0A182HZY3 A0A182XGE4 A0A182VFH3 A0A182JWC2 A0A182M141 Q380S9 A0A2S2NWJ8 A0A182RIV6 E2A055 A0A182VRH0 J9K946 A0A0J7L5L0
F4X0K5 A0A195F7N3 A0A0C9QPY2 A0A3S2N4Q7 A0A0C9QCN7 A0A1E1W253 E2B6U5 A0A194Q131 A0A1B6HSN4 A0A1B6GNX9 A0A0N1IAB6 A0A2A3EJP6 A0A088AGA7 A0A212F2Q5 A0A026WGX4 A0A154PMK4 A0A0M9ACA5 D6WBN5 A0A0P5A9Q3 A0A0P5DP87 U5EQE7 A0A2P8YTM3 A0A1Y1LAC1 A0A1Y1LF29 A0A1Y1LA95 A0A1Y1LG56 A0A0N8EMW7 A0A1L8DYB1 Q17P93 A0A1L8DYS0 A0A0P6ADJ6 A0A1B0CEV6 B0W6P5 A0A023ET90 A0A2M4BK03 A0A2M4BK52 A0A2M4BK12 A0A2M4BJY5 A0A182J1V5 A0A336K4K1 A0A1B6ECR7 V5GTI7 B4QBI8 A0A232FLL4 A0A0B4LGJ9 W8C971 A0A1I8M244 A0A1E1VYY8 B4I2C7 B4GCJ6 A0A182FRW8 W8BEE7 A0A1I8Q8R5 A0A1I8M243 A0A2H1VUV3 A0A1L8EDT9 A0A0L0CDG6 E0VSI7 A0A1L8EDX2 A0A1I8Q8N5 W5J2T7 A0A3B0JKR7 T1P9H1 A0A1A9XAC6 A0A2M4CZG5 A0A1L8EIS8 A0A1A9ZVJ6 A0A2M4ACX9 A0A1A9VKM6 A0A1B0FK30 A0A2M3Z840 A0A1I8Q8N3 A0A2M3Z7V7 A0A182TYJ3 A0A1W4V042 A0A182QDW4 A0A1B0C1H5 A0A2S2QGS3 A0A1B6JYC1 A0A182XW00 A0A182LR17 B4J6E5 A0A182HZY3 A0A182XGE4 A0A182VFH3 A0A182JWC2 A0A182M141 Q380S9 A0A2S2NWJ8 A0A182RIV6 E2A055 A0A182VRH0 J9K946 A0A0J7L5L0
Pubmed
19121390
20075255
21347285
21719571
20798317
26354079
+ More
22118469 24508170 18362917 19820115 29403074 28004739 17510324 24945155 17994087 22936249 28648823 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 24495485 25315136 26108605 20566863 20920257 23761445 25244985 20966253 12364791 14747013 17210077
22118469 24508170 18362917 19820115 29403074 28004739 17510324 24945155 17994087 22936249 28648823 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 24495485 25315136 26108605 20566863 20920257 23761445 25244985 20966253 12364791 14747013 17210077
EMBL
BABH01031817
BABH01031818
AAZX01006711
KQ981135
KYN09248.1
KQ977012
+ More
KYN06403.1 ADTU01012182 ADTU01012183 KQ976558 KYM80664.1 GL888498 EGI59997.1 KQ981744 KYN36391.1 GBYB01005709 JAG75476.1 RSAL01000568 RVE41378.1 GBYB01012183 JAG81950.1 GDQN01010040 JAT81014.1 GL446009 EFN88586.1 KQ459580 KPI99261.1 GECU01030036 JAS77670.1 GECZ01005635 JAS64134.1 KQ460396 KPJ15331.1 KZ288235 PBC31406.1 AGBW02010693 OWR48025.1 KK107213 EZA55285.1 KQ434980 KZC13082.1 KQ435706 KOX80219.1 KQ971311 EEZ98897.2 GDIP01205541 JAJ17861.1 GDIP01153636 LRGB01001581 JAJ69766.1 KZS11164.1 GANO01003300 JAB56571.1 PYGN01000366 PSN47586.1 GEZM01061279 JAV70574.1 GEZM01061278 JAV70575.1 GEZM01061280 JAV70572.1 GEZM01061277 JAV70576.1 GDIQ01011132 JAN83605.1 GFDF01002627 JAV11457.1 CH477193 EAT48539.1 GFDF01002642 JAV11442.1 GDIP01031146 JAM72569.1 AJWK01009303 AJWK01009304 AJWK01009305 AJWK01009306 AJWK01009307 DS231849 EDS36876.1 GAPW01001058 JAC12540.1 GGFJ01004244 MBW53385.1 GGFJ01004243 MBW53384.1 GGFJ01004246 MBW53387.1 GGFJ01004245 MBW53386.1 AXCP01008788 UFQS01000098 UFQT01000098 SSW99437.1 SSX19817.1 GEDC01001611 JAS35687.1 GALX01004913 JAB63553.1 CM000362 CM002911 EDX08506.1 KMY96249.1 NNAY01000061 OXU31399.1 AE013599 AHN56613.1 GAMC01006896 JAB99659.1 GDQN01011160 JAT79894.1 CH480820 EDW53922.1 CH479181 EDW32473.1 GAMC01006895 JAB99660.1 ODYU01004406 SOQ44242.1 GFDG01001932 JAV16867.1 JRES01000655 KNC29514.1 DS235751 EEB16343.1 GFDG01001933 JAV16866.1 ADMH02002129 ETN58607.1 OUUW01000001 SPP73231.1 KA644810 AFP59439.1 GGFL01006538 MBW70716.1 GFDG01000192 JAV18607.1 GGFK01005325 MBW38646.1 CCAG010017210 GGFM01003857 MBW24608.1 GGFM01003851 MBW24602.1 AXCN02001179 JXJN01024053 GGMS01007735 MBY76938.1 GECU01003502 JAT04205.1 CH916367 EDW01946.1 APCN01002019 AXCM01007505 AAAB01008987 EAL38481.4 GGMR01008885 MBY21504.1 GL435551 EFN73188.1 ABLF02011624 LBMM01000472 KMQ98257.1
KYN06403.1 ADTU01012182 ADTU01012183 KQ976558 KYM80664.1 GL888498 EGI59997.1 KQ981744 KYN36391.1 GBYB01005709 JAG75476.1 RSAL01000568 RVE41378.1 GBYB01012183 JAG81950.1 GDQN01010040 JAT81014.1 GL446009 EFN88586.1 KQ459580 KPI99261.1 GECU01030036 JAS77670.1 GECZ01005635 JAS64134.1 KQ460396 KPJ15331.1 KZ288235 PBC31406.1 AGBW02010693 OWR48025.1 KK107213 EZA55285.1 KQ434980 KZC13082.1 KQ435706 KOX80219.1 KQ971311 EEZ98897.2 GDIP01205541 JAJ17861.1 GDIP01153636 LRGB01001581 JAJ69766.1 KZS11164.1 GANO01003300 JAB56571.1 PYGN01000366 PSN47586.1 GEZM01061279 JAV70574.1 GEZM01061278 JAV70575.1 GEZM01061280 JAV70572.1 GEZM01061277 JAV70576.1 GDIQ01011132 JAN83605.1 GFDF01002627 JAV11457.1 CH477193 EAT48539.1 GFDF01002642 JAV11442.1 GDIP01031146 JAM72569.1 AJWK01009303 AJWK01009304 AJWK01009305 AJWK01009306 AJWK01009307 DS231849 EDS36876.1 GAPW01001058 JAC12540.1 GGFJ01004244 MBW53385.1 GGFJ01004243 MBW53384.1 GGFJ01004246 MBW53387.1 GGFJ01004245 MBW53386.1 AXCP01008788 UFQS01000098 UFQT01000098 SSW99437.1 SSX19817.1 GEDC01001611 JAS35687.1 GALX01004913 JAB63553.1 CM000362 CM002911 EDX08506.1 KMY96249.1 NNAY01000061 OXU31399.1 AE013599 AHN56613.1 GAMC01006896 JAB99659.1 GDQN01011160 JAT79894.1 CH480820 EDW53922.1 CH479181 EDW32473.1 GAMC01006895 JAB99660.1 ODYU01004406 SOQ44242.1 GFDG01001932 JAV16867.1 JRES01000655 KNC29514.1 DS235751 EEB16343.1 GFDG01001933 JAV16866.1 ADMH02002129 ETN58607.1 OUUW01000001 SPP73231.1 KA644810 AFP59439.1 GGFL01006538 MBW70716.1 GFDG01000192 JAV18607.1 GGFK01005325 MBW38646.1 CCAG010017210 GGFM01003857 MBW24608.1 GGFM01003851 MBW24602.1 AXCN02001179 JXJN01024053 GGMS01007735 MBY76938.1 GECU01003502 JAT04205.1 CH916367 EDW01946.1 APCN01002019 AXCM01007505 AAAB01008987 EAL38481.4 GGMR01008885 MBY21504.1 GL435551 EFN73188.1 ABLF02011624 LBMM01000472 KMQ98257.1
Proteomes
UP000005204
UP000002358
UP000078492
UP000078542
UP000005205
UP000078540
+ More
UP000007755 UP000078541 UP000283053 UP000008237 UP000053268 UP000053240 UP000242457 UP000005203 UP000007151 UP000053097 UP000076502 UP000053105 UP000007266 UP000076858 UP000245037 UP000008820 UP000092461 UP000002320 UP000075880 UP000000304 UP000215335 UP000000803 UP000095301 UP000001292 UP000008744 UP000069272 UP000095300 UP000037069 UP000009046 UP000000673 UP000268350 UP000092443 UP000092445 UP000078200 UP000092444 UP000075902 UP000192221 UP000075886 UP000092460 UP000076408 UP000075882 UP000001070 UP000075840 UP000076407 UP000075903 UP000075881 UP000075883 UP000007062 UP000075900 UP000000311 UP000075920 UP000007819 UP000036403
UP000007755 UP000078541 UP000283053 UP000008237 UP000053268 UP000053240 UP000242457 UP000005203 UP000007151 UP000053097 UP000076502 UP000053105 UP000007266 UP000076858 UP000245037 UP000008820 UP000092461 UP000002320 UP000075880 UP000000304 UP000215335 UP000000803 UP000095301 UP000001292 UP000008744 UP000069272 UP000095300 UP000037069 UP000009046 UP000000673 UP000268350 UP000092443 UP000092445 UP000078200 UP000092444 UP000075902 UP000192221 UP000075886 UP000092460 UP000076408 UP000075882 UP000001070 UP000075840 UP000076407 UP000075903 UP000075881 UP000075883 UP000007062 UP000075900 UP000000311 UP000075920 UP000007819 UP000036403
PRIDE
Pfam
PF01553 Acyltransferase
Interpro
IPR002123
Plipid/glycerol_acylTrfase
ProteinModelPortal
H9JRN3
K7IT41
A0A195D8P5
A0A195D0G0
A0A158ND16
A0A195B8Y9
+ More
F4X0K5 A0A195F7N3 A0A0C9QPY2 A0A3S2N4Q7 A0A0C9QCN7 A0A1E1W253 E2B6U5 A0A194Q131 A0A1B6HSN4 A0A1B6GNX9 A0A0N1IAB6 A0A2A3EJP6 A0A088AGA7 A0A212F2Q5 A0A026WGX4 A0A154PMK4 A0A0M9ACA5 D6WBN5 A0A0P5A9Q3 A0A0P5DP87 U5EQE7 A0A2P8YTM3 A0A1Y1LAC1 A0A1Y1LF29 A0A1Y1LA95 A0A1Y1LG56 A0A0N8EMW7 A0A1L8DYB1 Q17P93 A0A1L8DYS0 A0A0P6ADJ6 A0A1B0CEV6 B0W6P5 A0A023ET90 A0A2M4BK03 A0A2M4BK52 A0A2M4BK12 A0A2M4BJY5 A0A182J1V5 A0A336K4K1 A0A1B6ECR7 V5GTI7 B4QBI8 A0A232FLL4 A0A0B4LGJ9 W8C971 A0A1I8M244 A0A1E1VYY8 B4I2C7 B4GCJ6 A0A182FRW8 W8BEE7 A0A1I8Q8R5 A0A1I8M243 A0A2H1VUV3 A0A1L8EDT9 A0A0L0CDG6 E0VSI7 A0A1L8EDX2 A0A1I8Q8N5 W5J2T7 A0A3B0JKR7 T1P9H1 A0A1A9XAC6 A0A2M4CZG5 A0A1L8EIS8 A0A1A9ZVJ6 A0A2M4ACX9 A0A1A9VKM6 A0A1B0FK30 A0A2M3Z840 A0A1I8Q8N3 A0A2M3Z7V7 A0A182TYJ3 A0A1W4V042 A0A182QDW4 A0A1B0C1H5 A0A2S2QGS3 A0A1B6JYC1 A0A182XW00 A0A182LR17 B4J6E5 A0A182HZY3 A0A182XGE4 A0A182VFH3 A0A182JWC2 A0A182M141 Q380S9 A0A2S2NWJ8 A0A182RIV6 E2A055 A0A182VRH0 J9K946 A0A0J7L5L0
F4X0K5 A0A195F7N3 A0A0C9QPY2 A0A3S2N4Q7 A0A0C9QCN7 A0A1E1W253 E2B6U5 A0A194Q131 A0A1B6HSN4 A0A1B6GNX9 A0A0N1IAB6 A0A2A3EJP6 A0A088AGA7 A0A212F2Q5 A0A026WGX4 A0A154PMK4 A0A0M9ACA5 D6WBN5 A0A0P5A9Q3 A0A0P5DP87 U5EQE7 A0A2P8YTM3 A0A1Y1LAC1 A0A1Y1LF29 A0A1Y1LA95 A0A1Y1LG56 A0A0N8EMW7 A0A1L8DYB1 Q17P93 A0A1L8DYS0 A0A0P6ADJ6 A0A1B0CEV6 B0W6P5 A0A023ET90 A0A2M4BK03 A0A2M4BK52 A0A2M4BK12 A0A2M4BJY5 A0A182J1V5 A0A336K4K1 A0A1B6ECR7 V5GTI7 B4QBI8 A0A232FLL4 A0A0B4LGJ9 W8C971 A0A1I8M244 A0A1E1VYY8 B4I2C7 B4GCJ6 A0A182FRW8 W8BEE7 A0A1I8Q8R5 A0A1I8M243 A0A2H1VUV3 A0A1L8EDT9 A0A0L0CDG6 E0VSI7 A0A1L8EDX2 A0A1I8Q8N5 W5J2T7 A0A3B0JKR7 T1P9H1 A0A1A9XAC6 A0A2M4CZG5 A0A1L8EIS8 A0A1A9ZVJ6 A0A2M4ACX9 A0A1A9VKM6 A0A1B0FK30 A0A2M3Z840 A0A1I8Q8N3 A0A2M3Z7V7 A0A182TYJ3 A0A1W4V042 A0A182QDW4 A0A1B0C1H5 A0A2S2QGS3 A0A1B6JYC1 A0A182XW00 A0A182LR17 B4J6E5 A0A182HZY3 A0A182XGE4 A0A182VFH3 A0A182JWC2 A0A182M141 Q380S9 A0A2S2NWJ8 A0A182RIV6 E2A055 A0A182VRH0 J9K946 A0A0J7L5L0
Ontologies
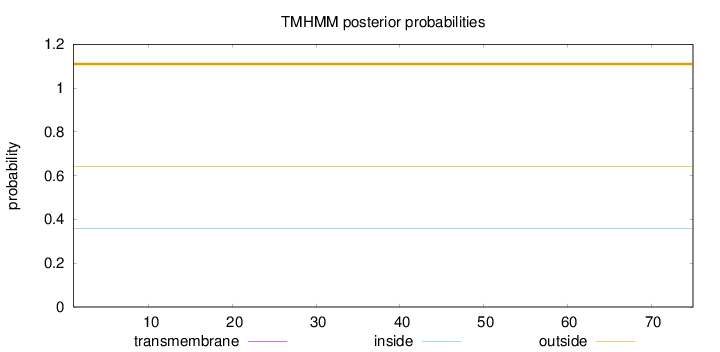
Topology
Length:
75
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00119
Exp number, first 60 AAs:
0.00119
Total prob of N-in:
0.35786
outside
1 - 75
Population Genetic Test Statistics
Pi
142.383437
Theta
135.144598
Tajima's D
0.468005
CLR
0.000085
CSRT
0.505024748762562
Interpretation
Uncertain
