Gene
KWMTBOMO14845 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012151
Annotation
ARP1_actin-related_protein_1-like_protein_A_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.64
Sequence
CDS
ATGGAGTTGAGTGATGTTATAGTGAATCAGCCCGTCGTAATAGACAACGGCTCCGGCGTAATAAAGGCGGGCTTCGCTGGTGATCAGATACCGAAATGTCGTTTCCCGAACTATATTGGTCGTCCGAAGCACACAAGAGTGATGGCAGGAGCGCTAGAAGGCGAGATCTTCGTTGGTCCTCGTGCAGAAGAGCATCGCGGCCTTCTCAGCATCAAATACCCAATGGAACACGGAATAGTTACAGACTGGAACGACATGGAAAAGATTTGGAATTATATTTACAGTAAGGACCAGCTATCTACTTTCTCAGAAGAACACCCAGTGCTGTTGACAGAAGCACCTCTGAACCCTCGCAGGAATAGAGAAAAGTCCGCCGAAATATTCTTTGAGACATTCAATGTACCGGCATTATTTCTTTCAATGCAGGCCGTTCTCAGTTTGTATGCAACAGGTCGTACCACGGGTGCTGTACTGGACTCCGGCGATGGAGTGACGCACGCCGTCCCCATATACGAGGGGTTCGCGATGCCGCACAGCATCATGAGAGTCGACGTGGCCGGGCGTGACGTCACCAAGTACCTTAGACTTCTACTCCGCAAAGAGGGAGTGAACCTCAGGACGTCCGCCGAGCTGGAGATCGTCAAGTCTATCAAGGAGCGGGCGTGCTACCTGTCTCCGAACCCTCTGAAGGAAGAGACCTTGGAGAATGAAAGGGCCCAATACGTTCTCCCTGATGGAACACAACTTGAGATTGGCCCGGCCCGGTTCCGAGCTCCCGAAGTGCTCTTCAGACCGGACCTGATCGGTGAGGAGTGCGAAGGTCTTCACGAGGTGTTAATGTTTTCAATCCACAAGTCGGACATGGACCTAAGGAAGGTCCTCTACCAGAACATAGTCCTCAGCGGGGGCTCCACGCTGTTCCGGGGGTTCGGGGACAGGCTCCTGCACGAGATCCGGAGACTAGCGCCTAAGGATATGAAGATCAGGATCTCGGCCCCCCAGGAGCGCCTGTACTCCACGTGGATCGGAGGCTCGATCCTCGCGTCTCTGGACACTTTCCGCAAGATGTGGGTCTCCAAGAGAGAGTATGACGAGGAGGGCCACAGGGCCGTCCACAGAAAGACGTTCTAG
Protein
MELSDVIVNQPVVIDNGSGVIKAGFAGDQIPKCRFPNYIGRPKHTRVMAGALEGEIFVGPRAEEHRGLLSIKYPMEHGIVTDWNDMEKIWNYIYSKDQLSTFSEEHPVLLTEAPLNPRRNREKSAEIFFETFNVPALFLSMQAVLSLYATGRTTGAVLDSGDGVTHAVPIYEGFAMPHSIMRVDVAGRDVTKYLRLLLRKEGVNLRTSAELEIVKSIKERACYLSPNPLKEETLENERAQYVLPDGTQLEIGPARFRAPEVLFRPDLIGEECEGLHEVLMFSIHKSDMDLRKVLYQNIVLSGGSTLFRGFGDRLLHEIRRLAPKDMKIRISAPQERLYSTWIGGSILASLDTFRKMWVSKREYDEEGHRAVHRKTF
Summary
Similarity
Belongs to the actin family.
Uniprot
Q1HQC8
A0A1E1WRB9
A0A2W1BIG3
A0A194Q1Q1
A0A212F2S0
A0A067QRR9
+ More
A0A2J7RH58 A0A2P8XLQ5 A0A2A3EF48 A0A0C9QZ95 E2A6V0 A0A158NKI3 A0A3L8DSF3 A0A088ADG4 C3Y5C1 E2C314 A0A1B6GR95 C1BUD7 A0A1B6E5Q5 A0A0K8TTV7 A0A1W4X687 A0A2R5LKE3 V5G2Y5 A0A0A9W7L5 T1ISG9 A0A1B0EZF3 E0VYK5 A0A293LWR2 A0A1L8E4V0 A0A1S3DBU9 A0A0P4VSM7 A0A069DYT4 A0A0V0G8M1 A0A023FJF1 A0A023FW29 M7BLL8 A0A023GJC8 G3MLF0 B7QDE8 A0A151NUW4 Q6DJH8 A9UMK9 A0A224YVH0 A0A131YWK4 L7MB37 A0A3L8SHE4 U3K221 V9KE22 A0A2P6KJ71 A0A3M6V2U3 A0A131XKV4 A0A226MSL1 A0A226PSC1 Q5ZM58 A0A0Q3T9Y1 L5K097 H3AQ73 A0A2B4SVY6 A0A1D2NMV2 A0A091VRB5 A0A0M8ZZJ5 A0A1Y1N7T7 W5N4M8 K1Q811 A0A1W7RJ85 H3B8R3 J3SE10 T1DMC6 A0A2D4HBT3 H0WTX1 A0A1Z5L798 A0A2K6F0Y1 A0A3B0K413 Q299V2 B4G5S4 A0A1V4KIK2 A0A2I0MEQ5 A0A1S3F9Q5 A0A1A6GV67 I3MMX5 A0A1U8BI71 H0VSV0 A0A091DPY8 G3HXW9 G5CAH6 A0A2K6BCH4 A0A2K6LIG1 A0A384DNN8 G1SPM5 G1P6V4 G7PDV1 G3QJD6 G1MHH5 G1RYT8 L8J3G7 A0A341AL98 A0A2K5ZZV9 A0A0D9QZD8 G3SVH1 A0A2Y9D8T9
A0A2J7RH58 A0A2P8XLQ5 A0A2A3EF48 A0A0C9QZ95 E2A6V0 A0A158NKI3 A0A3L8DSF3 A0A088ADG4 C3Y5C1 E2C314 A0A1B6GR95 C1BUD7 A0A1B6E5Q5 A0A0K8TTV7 A0A1W4X687 A0A2R5LKE3 V5G2Y5 A0A0A9W7L5 T1ISG9 A0A1B0EZF3 E0VYK5 A0A293LWR2 A0A1L8E4V0 A0A1S3DBU9 A0A0P4VSM7 A0A069DYT4 A0A0V0G8M1 A0A023FJF1 A0A023FW29 M7BLL8 A0A023GJC8 G3MLF0 B7QDE8 A0A151NUW4 Q6DJH8 A9UMK9 A0A224YVH0 A0A131YWK4 L7MB37 A0A3L8SHE4 U3K221 V9KE22 A0A2P6KJ71 A0A3M6V2U3 A0A131XKV4 A0A226MSL1 A0A226PSC1 Q5ZM58 A0A0Q3T9Y1 L5K097 H3AQ73 A0A2B4SVY6 A0A1D2NMV2 A0A091VRB5 A0A0M8ZZJ5 A0A1Y1N7T7 W5N4M8 K1Q811 A0A1W7RJ85 H3B8R3 J3SE10 T1DMC6 A0A2D4HBT3 H0WTX1 A0A1Z5L798 A0A2K6F0Y1 A0A3B0K413 Q299V2 B4G5S4 A0A1V4KIK2 A0A2I0MEQ5 A0A1S3F9Q5 A0A1A6GV67 I3MMX5 A0A1U8BI71 H0VSV0 A0A091DPY8 G3HXW9 G5CAH6 A0A2K6BCH4 A0A2K6LIG1 A0A384DNN8 G1SPM5 G1P6V4 G7PDV1 G3QJD6 G1MHH5 G1RYT8 L8J3G7 A0A341AL98 A0A2K5ZZV9 A0A0D9QZD8 G3SVH1 A0A2Y9D8T9
Pubmed
19121390
28756777
26354079
22118469
24845553
29403074
+ More
20798317 21347285 30249741 18563158 26369729 25401762 20566863 27129103 26334808 23624526 22216098 22293439 27762356 20431018 28797301 26830274 25576852 30282656 24402279 30382153 28049606 24621616 15592404 15642098 23258410 9215903 27289101 28004739 22992520 26358130 23025625 23758969 25727380 28528879 15632085 17994087 23371554 26319212 21993624 21804562 23929341 21993625 24813606 22002653 22398555 20010809 22751099
20798317 21347285 30249741 18563158 26369729 25401762 20566863 27129103 26334808 23624526 22216098 22293439 27762356 20431018 28797301 26830274 25576852 30282656 24402279 30382153 28049606 24621616 15592404 15642098 23258410 9215903 27289101 28004739 22992520 26358130 23025625 23758969 25727380 28528879 15632085 17994087 23371554 26319212 21993624 21804562 23929341 21993625 24813606 22002653 22398555 20010809 22751099
EMBL
BABH01031823
DQ443124
ABF51213.1
GDQN01001526
JAT89528.1
KZ150102
+ More
PZC73495.1 KQ459580 KPI99253.1 AGBW02010693 OWR48022.1 KK853097 KDR11422.1 NEVH01003746 PNF40171.1 PYGN01001763 PSN32946.1 KZ288271 PBC29829.1 GBYB01006027 JAG75794.1 GL437203 EFN70816.1 ADTU01002067 QOIP01000005 RLU23103.1 GG666487 EEN64168.1 GL452257 EFN77671.1 GECZ01004849 JAS64920.1 BT078216 HACA01013719 ACO12640.1 CDW31080.1 GEDC01004028 JAS33270.1 GDAI01000032 JAI17571.1 GGLE01005856 MBY09982.1 GALX01004086 JAB64380.1 GBHO01039172 GBHO01039169 GBHO01039168 GBHO01039166 JAG04432.1 JAG04435.1 JAG04436.1 JAG04438.1 JH431432 AJWK01022428 DS235845 EEB18461.1 GFWV01008434 MAA33163.1 GFDF01000316 JAV13768.1 GDKW01001009 JAI55586.1 GBGD01001835 JAC87054.1 GECL01001750 JAP04374.1 GBBK01002720 JAC21762.1 GBBL01001294 JAC26026.1 KB559124 EMP29092.1 GBBM01001404 JAC34014.1 JO842701 AEO34318.1 ABJB010284674 ABJB010736588 ABJB011057356 DS913072 EEC16870.1 AKHW03001922 KYO40646.1 BC075201 CM004478 AAH75201.1 OCT71662.1 AAMC01084081 BC157698 AAI57699.1 GFPF01006686 MAA17832.1 GEDV01005220 JAP83337.1 GACK01004670 JAA60364.1 QUSF01000019 RLW02164.1 AGTO01020393 JW863474 AFO95991.1 MWRG01010659 PRD26386.1 RCHS01000204 RMX60273.1 GEFH01000447 JAP68134.1 MCFN01000481 OXB58277.1 AWGT02000026 OXB82400.1 AADN05000306 AJ719526 CAG31185.1 LMAW01002767 KQK77342.1 KB031059 ELK05000.1 AFYH01182352 AFYH01182353 AFYH01182354 AFYH01182355 AFYH01182356 AFYH01182357 AFYH01182358 AFYH01182359 AFYH01182360 AFYH01182361 LSMT01000021 PFX32585.1 LJIJ01000002 ODN06594.1 KL409842 KFQ92213.1 KQ435812 KOX72749.1 GEZM01015377 JAV91697.1 AHAT01023142 AHAT01023143 AHAT01023144 JH817876 EKC30038.1 GDAY02000200 JAV51213.1 AFYH01078463 AFYH01078464 AFYH01078465 AFYH01078466 AFYH01078467 AFYH01078468 AFYH01078469 AFYH01078470 JU173930 GBEX01000276 AFJ49456.1 JAI14284.1 GAAZ01000213 GBKC01000290 GBKD01000185 JAA97730.1 JAG45780.1 JAG47433.1 IACK01019560 IACK01019561 LAA69428.1 AAQR03152039 GFJQ02003795 JAW03175.1 OUUW01000005 SPP80376.1 CM000070 EAL27599.1 CH479179 EDW24940.1 LSYS01003057 OPJ84274.1 AKCR02000017 PKK28142.1 LZPO01068676 OBS69525.1 AGTP01072689 AAKN02016521 KN122182 KFO32320.1 JH000895 KE672975 EGV98261.1 ERE78843.1 JH174169 GEBF01000039 EHB18537.1 JAO03594.1 AAGW02060858 AAPE02031085 CM001284 EHH64983.1 CABD030075840 CABD030075841 CABD030075842 ACTA01105810 ADFV01163572 ADFV01163573 ADFV01163574 ADFV01163575 JH880389 ELR61972.1 AQIB01042558
PZC73495.1 KQ459580 KPI99253.1 AGBW02010693 OWR48022.1 KK853097 KDR11422.1 NEVH01003746 PNF40171.1 PYGN01001763 PSN32946.1 KZ288271 PBC29829.1 GBYB01006027 JAG75794.1 GL437203 EFN70816.1 ADTU01002067 QOIP01000005 RLU23103.1 GG666487 EEN64168.1 GL452257 EFN77671.1 GECZ01004849 JAS64920.1 BT078216 HACA01013719 ACO12640.1 CDW31080.1 GEDC01004028 JAS33270.1 GDAI01000032 JAI17571.1 GGLE01005856 MBY09982.1 GALX01004086 JAB64380.1 GBHO01039172 GBHO01039169 GBHO01039168 GBHO01039166 JAG04432.1 JAG04435.1 JAG04436.1 JAG04438.1 JH431432 AJWK01022428 DS235845 EEB18461.1 GFWV01008434 MAA33163.1 GFDF01000316 JAV13768.1 GDKW01001009 JAI55586.1 GBGD01001835 JAC87054.1 GECL01001750 JAP04374.1 GBBK01002720 JAC21762.1 GBBL01001294 JAC26026.1 KB559124 EMP29092.1 GBBM01001404 JAC34014.1 JO842701 AEO34318.1 ABJB010284674 ABJB010736588 ABJB011057356 DS913072 EEC16870.1 AKHW03001922 KYO40646.1 BC075201 CM004478 AAH75201.1 OCT71662.1 AAMC01084081 BC157698 AAI57699.1 GFPF01006686 MAA17832.1 GEDV01005220 JAP83337.1 GACK01004670 JAA60364.1 QUSF01000019 RLW02164.1 AGTO01020393 JW863474 AFO95991.1 MWRG01010659 PRD26386.1 RCHS01000204 RMX60273.1 GEFH01000447 JAP68134.1 MCFN01000481 OXB58277.1 AWGT02000026 OXB82400.1 AADN05000306 AJ719526 CAG31185.1 LMAW01002767 KQK77342.1 KB031059 ELK05000.1 AFYH01182352 AFYH01182353 AFYH01182354 AFYH01182355 AFYH01182356 AFYH01182357 AFYH01182358 AFYH01182359 AFYH01182360 AFYH01182361 LSMT01000021 PFX32585.1 LJIJ01000002 ODN06594.1 KL409842 KFQ92213.1 KQ435812 KOX72749.1 GEZM01015377 JAV91697.1 AHAT01023142 AHAT01023143 AHAT01023144 JH817876 EKC30038.1 GDAY02000200 JAV51213.1 AFYH01078463 AFYH01078464 AFYH01078465 AFYH01078466 AFYH01078467 AFYH01078468 AFYH01078469 AFYH01078470 JU173930 GBEX01000276 AFJ49456.1 JAI14284.1 GAAZ01000213 GBKC01000290 GBKD01000185 JAA97730.1 JAG45780.1 JAG47433.1 IACK01019560 IACK01019561 LAA69428.1 AAQR03152039 GFJQ02003795 JAW03175.1 OUUW01000005 SPP80376.1 CM000070 EAL27599.1 CH479179 EDW24940.1 LSYS01003057 OPJ84274.1 AKCR02000017 PKK28142.1 LZPO01068676 OBS69525.1 AGTP01072689 AAKN02016521 KN122182 KFO32320.1 JH000895 KE672975 EGV98261.1 ERE78843.1 JH174169 GEBF01000039 EHB18537.1 JAO03594.1 AAGW02060858 AAPE02031085 CM001284 EHH64983.1 CABD030075840 CABD030075841 CABD030075842 ACTA01105810 ADFV01163572 ADFV01163573 ADFV01163574 ADFV01163575 JH880389 ELR61972.1 AQIB01042558
Proteomes
UP000005204
UP000053268
UP000007151
UP000027135
UP000235965
UP000245037
+ More
UP000242457 UP000000311 UP000005205 UP000279307 UP000005203 UP000001554 UP000008237 UP000192223 UP000092461 UP000009046 UP000079169 UP000031443 UP000001555 UP000050525 UP000186698 UP000008143 UP000276834 UP000016665 UP000275408 UP000198323 UP000198419 UP000000539 UP000051836 UP000010552 UP000008672 UP000225706 UP000094527 UP000053283 UP000053105 UP000018468 UP000005408 UP000005225 UP000233160 UP000268350 UP000001819 UP000008744 UP000190648 UP000053872 UP000081671 UP000092124 UP000005215 UP000189706 UP000005447 UP000028990 UP000001075 UP000030759 UP000006813 UP000233120 UP000233180 UP000261680 UP000291021 UP000001811 UP000001074 UP000009130 UP000001519 UP000008912 UP000001073 UP000252040 UP000233140 UP000029965 UP000007646 UP000248480
UP000242457 UP000000311 UP000005205 UP000279307 UP000005203 UP000001554 UP000008237 UP000192223 UP000092461 UP000009046 UP000079169 UP000031443 UP000001555 UP000050525 UP000186698 UP000008143 UP000276834 UP000016665 UP000275408 UP000198323 UP000198419 UP000000539 UP000051836 UP000010552 UP000008672 UP000225706 UP000094527 UP000053283 UP000053105 UP000018468 UP000005408 UP000005225 UP000233160 UP000268350 UP000001819 UP000008744 UP000190648 UP000053872 UP000081671 UP000092124 UP000005215 UP000189706 UP000005447 UP000028990 UP000001075 UP000030759 UP000006813 UP000233120 UP000233180 UP000261680 UP000291021 UP000001811 UP000001074 UP000009130 UP000001519 UP000008912 UP000001073 UP000252040 UP000233140 UP000029965 UP000007646 UP000248480
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
Q1HQC8
A0A1E1WRB9
A0A2W1BIG3
A0A194Q1Q1
A0A212F2S0
A0A067QRR9
+ More
A0A2J7RH58 A0A2P8XLQ5 A0A2A3EF48 A0A0C9QZ95 E2A6V0 A0A158NKI3 A0A3L8DSF3 A0A088ADG4 C3Y5C1 E2C314 A0A1B6GR95 C1BUD7 A0A1B6E5Q5 A0A0K8TTV7 A0A1W4X687 A0A2R5LKE3 V5G2Y5 A0A0A9W7L5 T1ISG9 A0A1B0EZF3 E0VYK5 A0A293LWR2 A0A1L8E4V0 A0A1S3DBU9 A0A0P4VSM7 A0A069DYT4 A0A0V0G8M1 A0A023FJF1 A0A023FW29 M7BLL8 A0A023GJC8 G3MLF0 B7QDE8 A0A151NUW4 Q6DJH8 A9UMK9 A0A224YVH0 A0A131YWK4 L7MB37 A0A3L8SHE4 U3K221 V9KE22 A0A2P6KJ71 A0A3M6V2U3 A0A131XKV4 A0A226MSL1 A0A226PSC1 Q5ZM58 A0A0Q3T9Y1 L5K097 H3AQ73 A0A2B4SVY6 A0A1D2NMV2 A0A091VRB5 A0A0M8ZZJ5 A0A1Y1N7T7 W5N4M8 K1Q811 A0A1W7RJ85 H3B8R3 J3SE10 T1DMC6 A0A2D4HBT3 H0WTX1 A0A1Z5L798 A0A2K6F0Y1 A0A3B0K413 Q299V2 B4G5S4 A0A1V4KIK2 A0A2I0MEQ5 A0A1S3F9Q5 A0A1A6GV67 I3MMX5 A0A1U8BI71 H0VSV0 A0A091DPY8 G3HXW9 G5CAH6 A0A2K6BCH4 A0A2K6LIG1 A0A384DNN8 G1SPM5 G1P6V4 G7PDV1 G3QJD6 G1MHH5 G1RYT8 L8J3G7 A0A341AL98 A0A2K5ZZV9 A0A0D9QZD8 G3SVH1 A0A2Y9D8T9
A0A2J7RH58 A0A2P8XLQ5 A0A2A3EF48 A0A0C9QZ95 E2A6V0 A0A158NKI3 A0A3L8DSF3 A0A088ADG4 C3Y5C1 E2C314 A0A1B6GR95 C1BUD7 A0A1B6E5Q5 A0A0K8TTV7 A0A1W4X687 A0A2R5LKE3 V5G2Y5 A0A0A9W7L5 T1ISG9 A0A1B0EZF3 E0VYK5 A0A293LWR2 A0A1L8E4V0 A0A1S3DBU9 A0A0P4VSM7 A0A069DYT4 A0A0V0G8M1 A0A023FJF1 A0A023FW29 M7BLL8 A0A023GJC8 G3MLF0 B7QDE8 A0A151NUW4 Q6DJH8 A9UMK9 A0A224YVH0 A0A131YWK4 L7MB37 A0A3L8SHE4 U3K221 V9KE22 A0A2P6KJ71 A0A3M6V2U3 A0A131XKV4 A0A226MSL1 A0A226PSC1 Q5ZM58 A0A0Q3T9Y1 L5K097 H3AQ73 A0A2B4SVY6 A0A1D2NMV2 A0A091VRB5 A0A0M8ZZJ5 A0A1Y1N7T7 W5N4M8 K1Q811 A0A1W7RJ85 H3B8R3 J3SE10 T1DMC6 A0A2D4HBT3 H0WTX1 A0A1Z5L798 A0A2K6F0Y1 A0A3B0K413 Q299V2 B4G5S4 A0A1V4KIK2 A0A2I0MEQ5 A0A1S3F9Q5 A0A1A6GV67 I3MMX5 A0A1U8BI71 H0VSV0 A0A091DPY8 G3HXW9 G5CAH6 A0A2K6BCH4 A0A2K6LIG1 A0A384DNN8 G1SPM5 G1P6V4 G7PDV1 G3QJD6 G1MHH5 G1RYT8 L8J3G7 A0A341AL98 A0A2K5ZZV9 A0A0D9QZD8 G3SVH1 A0A2Y9D8T9
PDB
6F3A
E-value=0,
Score=1719
Ontologies
GO
Topology
Subcellular location
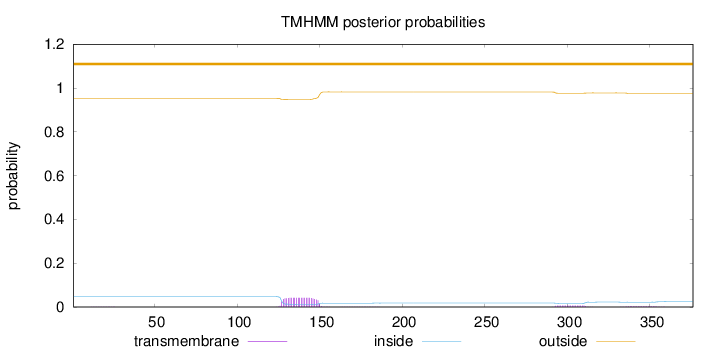
Length:
376
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.13915
Exp number, first 60 AAs:
0.0028
Total prob of N-in:
0.04820
outside
1 - 376
Population Genetic Test Statistics
Pi
224.268411
Theta
179.131809
Tajima's D
0.968878
CLR
0.604438
CSRT
0.647817609119544
Interpretation
Uncertain
