Gene
KWMTBOMO14844
Pre Gene Modal
BGIBMGA012194
Annotation
protein_O-fucosyltransferase_2_precursor_[Bombyx_mori]
Full name
GDP-fucose protein O-fucosyltransferase 2
Alternative Name
Peptide-O-fucosyltransferase 2
Location in the cell
Cytoplasmic Reliability : 1.674
Sequence
CDS
ATGAAACTAATTTCCACAATATTTTTTAATTTAATACTGATAAACGCTGCAATTAATACTGAAAATGGTTACTGTGTATCAAAGGACTCTTGTGCGGAAACAAAAAATACATCTTCCGACAAAAAATGGTTATTCTACGACGTTAATCCACCCGAAGGGTTTAATTTGCGACGAGACGTGTACATGAGATTCGCTATATTACTCTCCAACGAACAGAAACGAGGTAAACTGAAAGAGTTCGGATTAGTTCTACCGCCTTGGTATAAGCTGTACCATTGGAAAACCACAAAAAAGAGAGAACCAATTCCCTGGGGAAATTTTTTCGATATAGAAAGCTTAAAATCATTCGCGCCTGTCGTTGAGTTGTACGAGATATTCGAAAAATCTCTACAAAAGCAATTGATAATAGATACGATTTACGTGTTGCAAAACTACGACAATCCCTTCGAAAATGGCCTTTTCGAAGACAAATGGGAGGTACAGGAGGAATGTCCGTACGTGCCACAGTACTGGGGCTACGGAAACATAACGGCCAACGAGATAATCTGTGTTAAATTTCAAGGAAAAATTTCCATGCTACCGGAACTTTTTCAATTACACCGTAATGACAAATTCATAATGGTGGCCCACGGTGAAATACCTCTACACGATAATTATGGCGACCGAAAATACTGGAAATGCAGGAAAAGTATGAAATTCAATAAGAAACTAACAGAAATTGCTTTTAATTACATTAAAGAAAAATTAAATTGTGATACAGAGCTGTGCAAAAACTATTTAGGCGTGCACTGGAGGAGACAGGATTTTGCTAAATACAGGAGCAAAGACGTACCAACAACGTCTGGAACGGTCAAGCAAATCCAAAACATAATGCAAAACAAACTTCCTCATGTGCAAACAGTTTTCATAGCTACAGACGCATCGGCCAAAGAAAGAGAAATACTCCAGAATAATCTAGAAACTTTAGGTTATAACGTACATTATTATGTACCTAATATAGCTGAAGTTACCGAATTTGATGATGGTACTATTGCGATATTAGAACAAATAATTTGTTCACATGCTGCCTTCTTTGTGGGCACACATGAAAGTACCTTTACATTCCGAATACAAGAGGAAAGGGAGTTATTAGGGTTTCCTAGTGATACTACGTTCAATAGGCTGTGCCCCGACAAAGGCAATTGCGAAAAGCCATCCCGTTGGACTGTAGTGAATTAA
Protein
MKLISTIFFNLILINAAINTENGYCVSKDSCAETKNTSSDKKWLFYDVNPPEGFNLRRDVYMRFAILLSNEQKRGKLKEFGLVLPPWYKLYHWKTTKKREPIPWGNFFDIESLKSFAPVVELYEIFEKSLQKQLIIDTIYVLQNYDNPFENGLFEDKWEVQEECPYVPQYWGYGNITANEIICVKFQGKISMLPELFQLHRNDKFIMVAHGEIPLHDNYGDRKYWKCRKSMKFNKKLTEIAFNYIKEKLNCDTELCKNYLGVHWRRQDFAKYRSKDVPTTSGTVKQIQNIMQNKLPHVQTVFIATDASAKEREILQNNLETLGYNVHYYVPNIAEVTEFDDGTIAILEQIICSHAAFFVGTHESTFTFRIQEERELLGFPSDTTFNRLCPDKGNCEKPSRWTVVN
Summary
Description
Catalyzes the reaction that attaches fucose through an O-glycosidic linkage to a conserved serine or threonine residue in the consensus sequence C1-X(2,3)-S/T-C2-X(2)-G of thrombospondin type 1 repeats where C1 and C2 are the first and second cysteines, respectively. O-fucosylates members of several protein families including the ADAMTS family, the thrombosporin (TSP) and spondin families. The O-fucosylation of TSRs is also required for restricting epithelial to mesenchymal transition (EMT), maintaining the correct patterning of mesoderm and localization of the definite endoderm (By similarity). Required for the proper secretion of ADAMTS family members such as ADAMSL1 and ADAMST13.
Catalytic Activity
Transfers an alpha-L-fucosyl residue from GDP-beta-L-fucose to the serine hydroxy group of a protein acceptor.
Biophysicochemical Properties
9.8 uM for GDP-fucose
Similarity
Belongs to the glycosyltransferase 68 family.
Keywords
3D-structure
Alternative splicing
Carbohydrate metabolism
Complete proteome
Disulfide bond
Endoplasmic reticulum
Fucose metabolism
Glycoprotein
Glycosyltransferase
Golgi apparatus
Reference proteome
Signal
Transferase
Feature
chain GDP-fucose protein O-fucosyltransferase 2
splice variant In isoform B.
splice variant In isoform B.
Uniprot
Q00P36
Q659L5
A0A2A4KA84
A0A2W1BBV8
A0A0N0PDQ9
S4PXP5
+ More
A0A194Q0Z0 A0A212F2S7 A0A212F2Q4 D6WRI8 A0A1Y1MFF9 R4FP67 E0VEA7 A0A2P8XHD8 A0A1W4WSE4 J3JY99 A0A0P4VST8 A0A151WTF1 A0A158NRJ4 A0A154PIX3 E2C3D5 A0A151I8Y5 A0A026VZH5 A0A088ARB6 A0A146LL86 A0A0L7RCT4 A0A0A9WW19 A0A0P5PQ29 A0A0P5PMB5 A0A195BU85 A0A0N0BG72 A0A2P6KD75 A0A0P6FIZ7 A0A0P5RLY4 A0A182HBG0 V5GHC5 A0A1B6FEF3 V5II81 A0A2A3EBZ0 A0A224Z5R1 A0A0N8A3E4 A0A0P5VY32 A0A0P5B3V9 A0A0P6AIM2 A0A2C9JSB3 A0A131XSF2 L7M5C6 A0A164YLQ4 A0A3B3IBZ2 A0A3S1AQR3 A0A131YYT0 A0A3P9LMZ7 A0A0P5LST5 A0A1S4F6U9 Q0IFR1 C4WX22 A0A0P5AYQ8 A0A3P9H4A4 A0A0A1WW97 Q00P34 A0A3Q1B5L0 A0A3P8T7D1 A0A3Q2FYF9 A0A0P5AQP2 A0A2S2QIT1 A0A3B5K6P6 N6UR66 U4UB77 A0A0L0C8S0 F1L550 A0A067R730 A0A0P5R135 E9GXZ5 A0A0P5WUF9 A0A3Q1F6I5 A0A146NVL5 W8C4I8 A0A182IRA3 A0A3P8QBR5 A0A3B3B4N8 A0A3B4BE28 A0A2K5WB24 A0A087T4T4 K1P9F9 A0A3B4GWF5 A0A3Q2UU62 A0A3B3UQ17 A0A087X929 A0A2Y9E0H5 A0A2Y9G6Z9 I0FGL4 A0A182E546 A0A0S7H6B7 A0A044TK33 Q9Y2G5 A0A3Q2P4I9 G1QQA2 Q00P33 B0S641
A0A194Q0Z0 A0A212F2S7 A0A212F2Q4 D6WRI8 A0A1Y1MFF9 R4FP67 E0VEA7 A0A2P8XHD8 A0A1W4WSE4 J3JY99 A0A0P4VST8 A0A151WTF1 A0A158NRJ4 A0A154PIX3 E2C3D5 A0A151I8Y5 A0A026VZH5 A0A088ARB6 A0A146LL86 A0A0L7RCT4 A0A0A9WW19 A0A0P5PQ29 A0A0P5PMB5 A0A195BU85 A0A0N0BG72 A0A2P6KD75 A0A0P6FIZ7 A0A0P5RLY4 A0A182HBG0 V5GHC5 A0A1B6FEF3 V5II81 A0A2A3EBZ0 A0A224Z5R1 A0A0N8A3E4 A0A0P5VY32 A0A0P5B3V9 A0A0P6AIM2 A0A2C9JSB3 A0A131XSF2 L7M5C6 A0A164YLQ4 A0A3B3IBZ2 A0A3S1AQR3 A0A131YYT0 A0A3P9LMZ7 A0A0P5LST5 A0A1S4F6U9 Q0IFR1 C4WX22 A0A0P5AYQ8 A0A3P9H4A4 A0A0A1WW97 Q00P34 A0A3Q1B5L0 A0A3P8T7D1 A0A3Q2FYF9 A0A0P5AQP2 A0A2S2QIT1 A0A3B5K6P6 N6UR66 U4UB77 A0A0L0C8S0 F1L550 A0A067R730 A0A0P5R135 E9GXZ5 A0A0P5WUF9 A0A3Q1F6I5 A0A146NVL5 W8C4I8 A0A182IRA3 A0A3P8QBR5 A0A3B3B4N8 A0A3B4BE28 A0A2K5WB24 A0A087T4T4 K1P9F9 A0A3B4GWF5 A0A3Q2UU62 A0A3B3UQ17 A0A087X929 A0A2Y9E0H5 A0A2Y9G6Z9 I0FGL4 A0A182E546 A0A0S7H6B7 A0A044TK33 Q9Y2G5 A0A3Q2P4I9 G1QQA2 Q00P33 B0S641
EC Number
2.4.1.221
Pubmed
16679357
19121390
28756777
26354079
23622113
22118469
+ More
18362917 19820115 28004739 20566863 29403074 22516182 27129103 21347285 20798317 24508170 30249741 26823975 25401762 26483478 25765539 28797301 15562597 25576852 17554307 26830274 17510324 25830018 21551351 23537049 26108605 21685128 24845553 21292972 24495485 29451363 25463417 22992520 25319552 22919073 15233996 12966037 10231032 17974005 10830953 15489334 11067851 16464858 17395589 17395588 25944712 22588082 23594743
18362917 19820115 28004739 20566863 29403074 22516182 27129103 21347285 20798317 24508170 30249741 26823975 25401762 26483478 25765539 28797301 15562597 25576852 17554307 26830274 17510324 25830018 21551351 23537049 26108605 21685128 24845553 21292972 24495485 29451363 25463417 22992520 25319552 22919073 15233996 12966037 10231032 17974005 10830953 15489334 11067851 16464858 17395589 17395588 25944712 22588082 23594743
EMBL
BABH01031823
DQ139953
ABA29474.1
AJ831834
CAH41977.1
NWSH01000015
+ More
PCG80824.1 KZ150242 PZC71901.1 KQ460240 KPJ16389.1 GAIX01004678 JAA87882.1 KQ459580 KPI99252.1 AGBW02010693 OWR48021.1 OWR48020.1 KQ971351 EFA07652.2 GEZM01035974 GEZM01035973 GEZM01035972 GEZM01035971 JAV83065.1 GAHY01001034 JAA76476.1 DS235088 EEB11713.1 PYGN01002122 PSN31421.1 BT128223 AEE63184.1 GDKW01001552 JAI55043.1 KQ982757 KYQ51113.1 ADTU01002058 ADTU01002059 KQ434931 KZC11772.1 GL452320 EFN77473.1 KQ978331 KYM95069.1 KK107538 QOIP01000005 EZA49132.1 RLU22922.1 GDHC01009911 JAQ08718.1 KQ414615 KOC68600.1 GBHO01030958 GBHO01030957 GBRD01007399 GBRD01007398 JAG12646.1 JAG12647.1 JAG58422.1 GDIQ01146128 JAL05598.1 GDIQ01147518 JAL04208.1 KQ976403 KYM92137.1 KQ435791 KOX74368.1 MWRG01015540 PRD24271.1 GDIQ01063664 JAN31073.1 GDIQ01098847 JAL52879.1 JXUM01124797 KQ566870 KXJ69771.1 GANP01014848 JAB69620.1 GECZ01021285 GECZ01020965 JAS48484.1 JAS48804.1 GANP01003647 JAB80821.1 KZ288289 PBC29293.1 GFPF01010456 MAA21602.1 GDIP01186385 JAJ37017.1 GDIP01093592 JAM10123.1 GDIP01190095 JAJ33307.1 GDIP01030105 JAM73610.1 GEFM01006661 JAP69135.1 GACK01006735 JAA58299.1 LRGB01000872 KZS15384.1 RQTK01001694 RUS69404.1 GEDV01004845 JAP83712.1 GDIQ01165773 JAK85952.1 CH477294 EAT44474.1 ABLF02038228 AK342404 BAH72442.1 GDIP01196548 JAJ26854.1 GBXI01010993 JAD03299.1 DQ139955 ABA29476.1 GDIP01195437 JAJ27965.1 GGMS01008400 MBY77603.1 APGK01019954 KB740150 ENN81252.1 KB632173 ERL89578.1 JRES01000754 KNC28671.1 JI171547 ADY45254.1 KK852657 KDR19215.1 GDIQ01109391 JAL42335.1 GL732574 EFX75507.1 GDIP01081549 JAM22166.1 GCES01150539 JAQ35783.1 GAMC01002282 JAC04274.1 AQIA01044232 AQIA01044233 KK113396 KFM60123.1 JH818836 EKC20412.1 AYCK01006445 AYCK01006446 JV043519 AFI33590.1 UYRW01000565 VDK68049.1 GBYX01445506 GBYX01445502 JAO35931.1 CMVM020000146 AJ302080 AJ302079 AY066015 AJ575591 AB023175 AL110285 AL163301 BC011044 BC064623 ADFV01100857 ADFV01100858 DQ139956 ABA29477.1 BX323586 BC163087 BC163097 AAI63087.1
PCG80824.1 KZ150242 PZC71901.1 KQ460240 KPJ16389.1 GAIX01004678 JAA87882.1 KQ459580 KPI99252.1 AGBW02010693 OWR48021.1 OWR48020.1 KQ971351 EFA07652.2 GEZM01035974 GEZM01035973 GEZM01035972 GEZM01035971 JAV83065.1 GAHY01001034 JAA76476.1 DS235088 EEB11713.1 PYGN01002122 PSN31421.1 BT128223 AEE63184.1 GDKW01001552 JAI55043.1 KQ982757 KYQ51113.1 ADTU01002058 ADTU01002059 KQ434931 KZC11772.1 GL452320 EFN77473.1 KQ978331 KYM95069.1 KK107538 QOIP01000005 EZA49132.1 RLU22922.1 GDHC01009911 JAQ08718.1 KQ414615 KOC68600.1 GBHO01030958 GBHO01030957 GBRD01007399 GBRD01007398 JAG12646.1 JAG12647.1 JAG58422.1 GDIQ01146128 JAL05598.1 GDIQ01147518 JAL04208.1 KQ976403 KYM92137.1 KQ435791 KOX74368.1 MWRG01015540 PRD24271.1 GDIQ01063664 JAN31073.1 GDIQ01098847 JAL52879.1 JXUM01124797 KQ566870 KXJ69771.1 GANP01014848 JAB69620.1 GECZ01021285 GECZ01020965 JAS48484.1 JAS48804.1 GANP01003647 JAB80821.1 KZ288289 PBC29293.1 GFPF01010456 MAA21602.1 GDIP01186385 JAJ37017.1 GDIP01093592 JAM10123.1 GDIP01190095 JAJ33307.1 GDIP01030105 JAM73610.1 GEFM01006661 JAP69135.1 GACK01006735 JAA58299.1 LRGB01000872 KZS15384.1 RQTK01001694 RUS69404.1 GEDV01004845 JAP83712.1 GDIQ01165773 JAK85952.1 CH477294 EAT44474.1 ABLF02038228 AK342404 BAH72442.1 GDIP01196548 JAJ26854.1 GBXI01010993 JAD03299.1 DQ139955 ABA29476.1 GDIP01195437 JAJ27965.1 GGMS01008400 MBY77603.1 APGK01019954 KB740150 ENN81252.1 KB632173 ERL89578.1 JRES01000754 KNC28671.1 JI171547 ADY45254.1 KK852657 KDR19215.1 GDIQ01109391 JAL42335.1 GL732574 EFX75507.1 GDIP01081549 JAM22166.1 GCES01150539 JAQ35783.1 GAMC01002282 JAC04274.1 AQIA01044232 AQIA01044233 KK113396 KFM60123.1 JH818836 EKC20412.1 AYCK01006445 AYCK01006446 JV043519 AFI33590.1 UYRW01000565 VDK68049.1 GBYX01445506 GBYX01445502 JAO35931.1 CMVM020000146 AJ302080 AJ302079 AY066015 AJ575591 AB023175 AL110285 AL163301 BC011044 BC064623 ADFV01100857 ADFV01100858 DQ139956 ABA29477.1 BX323586 BC163087 BC163097 AAI63087.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000007266
+ More
UP000009046 UP000245037 UP000192223 UP000075809 UP000005205 UP000076502 UP000008237 UP000078542 UP000053097 UP000279307 UP000005203 UP000053825 UP000078540 UP000053105 UP000069940 UP000249989 UP000242457 UP000076420 UP000076858 UP000001038 UP000271974 UP000265180 UP000008820 UP000007819 UP000265200 UP000257160 UP000265080 UP000265020 UP000005226 UP000019118 UP000030742 UP000037069 UP000027135 UP000000305 UP000257200 UP000075880 UP000265100 UP000261560 UP000261520 UP000233100 UP000054359 UP000005408 UP000261460 UP000264840 UP000261500 UP000028760 UP000248480 UP000248481 UP000077448 UP000271087 UP000024404 UP000005640 UP000265000 UP000001073 UP000000437
UP000009046 UP000245037 UP000192223 UP000075809 UP000005205 UP000076502 UP000008237 UP000078542 UP000053097 UP000279307 UP000005203 UP000053825 UP000078540 UP000053105 UP000069940 UP000249989 UP000242457 UP000076420 UP000076858 UP000001038 UP000271974 UP000265180 UP000008820 UP000007819 UP000265200 UP000257160 UP000265080 UP000265020 UP000005226 UP000019118 UP000030742 UP000037069 UP000027135 UP000000305 UP000257200 UP000075880 UP000265100 UP000261560 UP000261520 UP000233100 UP000054359 UP000005408 UP000261460 UP000264840 UP000261500 UP000028760 UP000248480 UP000248481 UP000077448 UP000271087 UP000024404 UP000005640 UP000265000 UP000001073 UP000000437
Pfam
PF10250 O-FucT
Interpro
IPR019378
GDP-Fuc_O-FucTrfase
ProteinModelPortal
Q00P36
Q659L5
A0A2A4KA84
A0A2W1BBV8
A0A0N0PDQ9
S4PXP5
+ More
A0A194Q0Z0 A0A212F2S7 A0A212F2Q4 D6WRI8 A0A1Y1MFF9 R4FP67 E0VEA7 A0A2P8XHD8 A0A1W4WSE4 J3JY99 A0A0P4VST8 A0A151WTF1 A0A158NRJ4 A0A154PIX3 E2C3D5 A0A151I8Y5 A0A026VZH5 A0A088ARB6 A0A146LL86 A0A0L7RCT4 A0A0A9WW19 A0A0P5PQ29 A0A0P5PMB5 A0A195BU85 A0A0N0BG72 A0A2P6KD75 A0A0P6FIZ7 A0A0P5RLY4 A0A182HBG0 V5GHC5 A0A1B6FEF3 V5II81 A0A2A3EBZ0 A0A224Z5R1 A0A0N8A3E4 A0A0P5VY32 A0A0P5B3V9 A0A0P6AIM2 A0A2C9JSB3 A0A131XSF2 L7M5C6 A0A164YLQ4 A0A3B3IBZ2 A0A3S1AQR3 A0A131YYT0 A0A3P9LMZ7 A0A0P5LST5 A0A1S4F6U9 Q0IFR1 C4WX22 A0A0P5AYQ8 A0A3P9H4A4 A0A0A1WW97 Q00P34 A0A3Q1B5L0 A0A3P8T7D1 A0A3Q2FYF9 A0A0P5AQP2 A0A2S2QIT1 A0A3B5K6P6 N6UR66 U4UB77 A0A0L0C8S0 F1L550 A0A067R730 A0A0P5R135 E9GXZ5 A0A0P5WUF9 A0A3Q1F6I5 A0A146NVL5 W8C4I8 A0A182IRA3 A0A3P8QBR5 A0A3B3B4N8 A0A3B4BE28 A0A2K5WB24 A0A087T4T4 K1P9F9 A0A3B4GWF5 A0A3Q2UU62 A0A3B3UQ17 A0A087X929 A0A2Y9E0H5 A0A2Y9G6Z9 I0FGL4 A0A182E546 A0A0S7H6B7 A0A044TK33 Q9Y2G5 A0A3Q2P4I9 G1QQA2 Q00P33 B0S641
A0A194Q0Z0 A0A212F2S7 A0A212F2Q4 D6WRI8 A0A1Y1MFF9 R4FP67 E0VEA7 A0A2P8XHD8 A0A1W4WSE4 J3JY99 A0A0P4VST8 A0A151WTF1 A0A158NRJ4 A0A154PIX3 E2C3D5 A0A151I8Y5 A0A026VZH5 A0A088ARB6 A0A146LL86 A0A0L7RCT4 A0A0A9WW19 A0A0P5PQ29 A0A0P5PMB5 A0A195BU85 A0A0N0BG72 A0A2P6KD75 A0A0P6FIZ7 A0A0P5RLY4 A0A182HBG0 V5GHC5 A0A1B6FEF3 V5II81 A0A2A3EBZ0 A0A224Z5R1 A0A0N8A3E4 A0A0P5VY32 A0A0P5B3V9 A0A0P6AIM2 A0A2C9JSB3 A0A131XSF2 L7M5C6 A0A164YLQ4 A0A3B3IBZ2 A0A3S1AQR3 A0A131YYT0 A0A3P9LMZ7 A0A0P5LST5 A0A1S4F6U9 Q0IFR1 C4WX22 A0A0P5AYQ8 A0A3P9H4A4 A0A0A1WW97 Q00P34 A0A3Q1B5L0 A0A3P8T7D1 A0A3Q2FYF9 A0A0P5AQP2 A0A2S2QIT1 A0A3B5K6P6 N6UR66 U4UB77 A0A0L0C8S0 F1L550 A0A067R730 A0A0P5R135 E9GXZ5 A0A0P5WUF9 A0A3Q1F6I5 A0A146NVL5 W8C4I8 A0A182IRA3 A0A3P8QBR5 A0A3B3B4N8 A0A3B4BE28 A0A2K5WB24 A0A087T4T4 K1P9F9 A0A3B4GWF5 A0A3Q2UU62 A0A3B3UQ17 A0A087X929 A0A2Y9E0H5 A0A2Y9G6Z9 I0FGL4 A0A182E546 A0A0S7H6B7 A0A044TK33 Q9Y2G5 A0A3Q2P4I9 G1QQA2 Q00P33 B0S641
PDB
4AP5
E-value=2.64832e-68,
Score=657
Ontologies
GO
Topology
Subcellular location
Endoplasmic reticulum
Golgi apparatus
Golgi apparatus
SignalP
Position: 1 - 16,
Likelihood: 0.952763
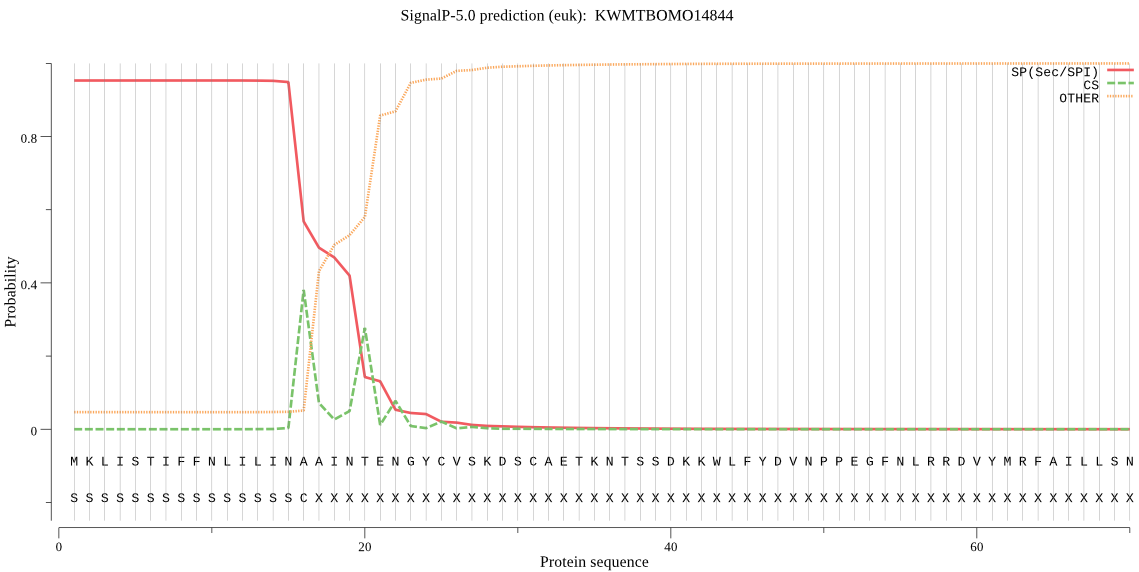
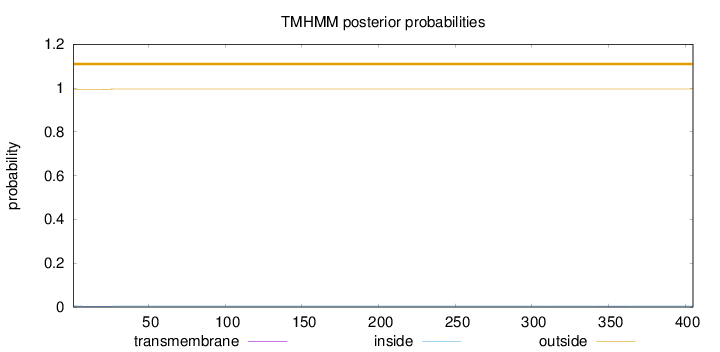
Length:
405
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0753699999999999
Exp number, first 60 AAs:
0.06792
Total prob of N-in:
0.00563
outside
1 - 405
Population Genetic Test Statistics
Pi
139.161835
Theta
148.92865
Tajima's D
-0.217927
CLR
67.890534
CSRT
0.310534473276336
Interpretation
Uncertain
