Pre Gene Modal
BGIBMGA012195
Annotation
PREDICTED:_double-stranded_RNA-specific_editase_Adar_isoform_X1_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.932 Nuclear Reliability : 1.809
Sequence
CDS
ATGAATATTACTTTTCAGACTTCGGTGTGCGATATAACGTCGACATCGGACTTTGTGAGTCCTCGAAGTAAGATGATCAAGCAGAAACGGAAACGCGAGTTTCCGTGTACAGATGTGCAACCTGCAATCAAGAGGCCCGCGCCATCGATACGTGTCGGCTCCGGATTCAAATGGAACCTACAGGCGTTGGCGCATGCGGCCGACGCGGTGCAGTCTGGCGGAAAATCACCTGTCTCCGCCCTCAACGAGCTGGGCGTTCGCGTCACCTACACCGTGCTGAGCCAGGGCGGCCCAGCGCATTGTCCCAGCTTCACCGTGGCAGTCAATCTTACCAAGAAGAAAAATAAGAGAACAATAAGTAAGATCTGTAAGGATTTAAATAAAAAGAATCCAGTGATGATATTGAATGAAACGCCACACGGATTTCGATGCAAGGAAATAGGAACCTTCAGTACTCCCAATAAGACGGTCTTTGGAGTTGCTATGACTGTCGCCGATATGACCTTTGAAGGTTGGGGTCACAGTAAGCGTGAGGCACGCGCGTCCGCAGCCAGGGTTTGTCTCGCGGCTCTGCTCGCCCGAGCTGGCCACGTGCTGCCAGACCAGACCCGCGCCAAGGACTTCACTGTTGACGACACAACACCTTATCCCGTCTCCGAATTCCAATGTCTCGAGAGCAAATCGAAGATTGAAGAGCAGACGAAGCAAAACGCTGTCACGGAAACAGCCCAGAGTCAGATTGAGAGGAATCTGCCCACGTCGTCGCTGTTCGGCGCCACCGCCCCCACCCCGGCCTCCAAGAGCCCCATCAATGTCCTCTACGAGAACCACCCGGGGCTGCTGTTCATCTGTACCTATGGGGACGGAACACCCTTGGATTCTGCTCAGGTTCCGCTACAGCTGGCCCACAGCATGAGGTTCAAGGTTGTCTGCCAGATCAAGGAACAGAAGTTCGAGGGTTATGGTTCGAGCAAGAAGCTGGCGAAGTTGGCGGCGGCCCGCAGCGCGCTGGCCGTGCTGGAGGAGGGCCGCGCCGACATCGCGGACAGCCTGCTGCCCTCCGGCTACCTCTCGCAGACCCTCGCCGACCACATAGCCAATCTCGTGAACAGCAAATTCAGTGAGATGATGAAGAATGACGTCATACATTCGAAGAGGAAGGTCCTGGCCGGGATCGTGATGACGACCGACAGCAAGGTGGACGGTGCCAAGGTGATAGCGGTGGCGACGGGCACCAAGTGCGTCTCCGGCGAGCACATGTCGGTGCGCGGCCGGGCCGTCAACGACTGCCACGCGGAGGTGGCGGCCCGGCGCTGCCTCCAACGCCACCTGTACACCCAGCTGCTGCTTTACGCCTCACAAAAAGACCCCCGCAAGCCGATCCCCGAGGCGGACATTGAGCCCGCGGAGGGGGGCGGCTACCGACTCCGCCCCGACCGGCAGCTACACATGTACGTGAGCACCGCGCCCTGCGGGGACGGGCGGATATTCAGCCCGCACGAGCAGCTACAGCACGAGCCGGACCGACATCCCAACAGGTTGGCCCGGGGACAGCTCCGCACGAAGATAGAGTCGGGCGAGGGTACGATCCCGGTGAAGAACTGCAACAGCTTGCTGCAGACCTGGGACGGTGTTATGCAGGGCGAGCGCCTCCTGACGATGTCGTGTTCTGATAAAGTGGCCCGCTGGTGCGTCCTCGGCCTGCAGGGCTCGCTGATGTCCACGCTGCTGGCGCCCGTGTACCCGACCTCGATAGTGCTGGGCTCGCTGCTGCACCCGCACCACTTGTACAGACCGAAAAATTTCTTGAAGAATCCATTCAACGCTGAACCTTTGCTCGCCTCTATATTATATGAGAAAGGCGTGCCAAACAACGCTGGTTTCGCGCCTATATAA
Protein
MNITFQTSVCDITSTSDFVSPRSKMIKQKRKREFPCTDVQPAIKRPAPSIRVGSGFKWNLQALAHAADAVQSGGKSPVSALNELGVRVTYTVLSQGGPAHCPSFTVAVNLTKKKNKRTISKICKDLNKKNPVMILNETPHGFRCKEIGTFSTPNKTVFGVAMTVADMTFEGWGHSKREARASAARVCLAALLARAGHVLPDQTRAKDFTVDDTTPYPVSEFQCLESKSKIEEQTKQNAVTETAQSQIERNLPTSSLFGATAPTPASKSPINVLYENHPGLLFICTYGDGTPLDSAQVPLQLAHSMRFKVVCQIKEQKFEGYGSSKKLAKLAAARSALAVLEEGRADIADSLLPSGYLSQTLADHIANLVNSKFSEMMKNDVIHSKRKVLAGIVMTTDSKVDGAKVIAVATGTKCVSGEHMSVRGRAVNDCHAEVAARRCLQRHLYTQLLLYASQKDPRKPIPEADIEPAEGGGYRLRPDRQLHMYVSTAPCGDGRIFSPHEQLQHEPDRHPNRLARGQLRTKIESGEGTIPVKNCNSLLQTWDGVMQGERLLTMSCSDKVARWCVLGLQGSLMSTLLAPVYPTSIVLGSLLHPHHLYRPKNFLKNPFNAEPLLASILYEKGVPNNAGFAPI
Summary
Uniprot
H9JRN5
A0A212F4F2
A0A194Q779
A0A091M2P3
A0A1D5PS41
K7FFP1
+ More
K7FFR2 A0A2K6SLH3 A0A2K6SLK0 A0A093HJK0 A0A091KMX0 V8PFX0 Q90VV7 Q90VZ5 A0A091HNZ3 A0A093DEP7 A0A091J7G2 A0A3Q0GE77 A0A1U7RXQ4 A0A093Q1Y1 A0A093FCI1 A0A2I3M1R9 A0A091V4A8 A0A093HW59 A0A099ZKD0 A0A151N9R2 A0A2K5CS14 A0A3B4Y9H6 A0A3B4U6H5 A0A2K5R875 A0A2K5CS27 A0A3Q0GEE6 A0A0A0AXY6 A0A091T6F8 U3DFM0 A0A087R9G8 A0A2K5R884 A0A093NXM0 A0A091Q284 U3J7Q1 A0A0S7L0C3 A0A1L8EWQ6 A0A091H2Q2 Q5U4T4 A0A3B4Y9J8 A0A2K5UG64 F7IS82 A0A096NIB1 A0A2Y9SBA4 A0A093GKP7 H0ZLN7 G1KJC4 A0A0S7L091 A0A2I3GLS2 F7FMP3 A0A3Q2Y3Q7 A4IIV9 A0A091V5K9 L5JX02 A0A218UNP1 A0A3P8WSX3 A0A3L8S3D5 S7Q3W8 A0A2K6BU96 A0A2K5UFW6 A0A0D9R561 A0A2K5LEP2 F7G963 A0A2R8ZY51 H2QZM5 A0A2K6BU98 A0A2K5UG24 A0A2I3LDH1 A0A2K5LEX0 A0A1D5QUE4 A0A2R8ZTH9 A0A2I3T8K9 A0A2I2V5I5 A0A2K6RDI1 A0A2K5HB13 G1QQ87 H2SWG8 A0A2K5HAZ8 A0A2J8RSE4 A0A2K6RDM5 A0A2J8RSD5 A0A1A8S8J9 A0A2K6L3P3 A0A091F477 A0A3Q4AR44 H0W0L7 A0A2K6RDG1 A0A3P8WQX3
K7FFR2 A0A2K6SLH3 A0A2K6SLK0 A0A093HJK0 A0A091KMX0 V8PFX0 Q90VV7 Q90VZ5 A0A091HNZ3 A0A093DEP7 A0A091J7G2 A0A3Q0GE77 A0A1U7RXQ4 A0A093Q1Y1 A0A093FCI1 A0A2I3M1R9 A0A091V4A8 A0A093HW59 A0A099ZKD0 A0A151N9R2 A0A2K5CS14 A0A3B4Y9H6 A0A3B4U6H5 A0A2K5R875 A0A2K5CS27 A0A3Q0GEE6 A0A0A0AXY6 A0A091T6F8 U3DFM0 A0A087R9G8 A0A2K5R884 A0A093NXM0 A0A091Q284 U3J7Q1 A0A0S7L0C3 A0A1L8EWQ6 A0A091H2Q2 Q5U4T4 A0A3B4Y9J8 A0A2K5UG64 F7IS82 A0A096NIB1 A0A2Y9SBA4 A0A093GKP7 H0ZLN7 G1KJC4 A0A0S7L091 A0A2I3GLS2 F7FMP3 A0A3Q2Y3Q7 A4IIV9 A0A091V5K9 L5JX02 A0A218UNP1 A0A3P8WSX3 A0A3L8S3D5 S7Q3W8 A0A2K6BU96 A0A2K5UFW6 A0A0D9R561 A0A2K5LEP2 F7G963 A0A2R8ZY51 H2QZM5 A0A2K6BU98 A0A2K5UG24 A0A2I3LDH1 A0A2K5LEX0 A0A1D5QUE4 A0A2R8ZTH9 A0A2I3T8K9 A0A2I2V5I5 A0A2K6RDI1 A0A2K5HB13 G1QQ87 H2SWG8 A0A2K5HAZ8 A0A2J8RSE4 A0A2K6RDM5 A0A2J8RSD5 A0A1A8S8J9 A0A2K6L3P3 A0A091F477 A0A3Q4AR44 H0W0L7 A0A2K6RDG1 A0A3P8WQX3
Pubmed
EMBL
BABH01031824
BABH01031825
BABH01031826
BABH01031827
BABH01031828
AGBW02010384
+ More
OWR48602.1 KQ459580 KPI99250.1 KK520006 KFP66068.1 AADN05000347 AGCU01020983 AGCU01020984 AGCU01020985 KL206176 KFV79635.1 KK534807 KFP29099.1 AZIM01000162 ETE72898.1 AF403117 AF403121 AAL01312.1 AF403115 AF403119 AJ719343 AAL01310.1 CAG31002.1 KL217603 KFO96884.1 KN125754 KFU84322.1 KK501508 KFP15670.1 KL670089 KFW78257.1 KK630484 KFV55417.1 AHZZ02021396 AHZZ02021397 KL410606 KFQ97843.1 KK398741 KFV58723.1 KL895122 KGL82296.1 AKHW03003682 KYO33572.1 KL873342 KGL98378.1 KK438364 KFQ69425.1 GAMS01000760 GAMR01004025 GAMQ01004428 JAB22376.1 JAB29907.1 JAB37423.1 KL226224 KFM10122.1 KL224844 KFW64847.1 KK683546 KFQ13626.1 ADON01053263 GBYX01190734 JAO82930.1 CM004482 OCT63787.1 KL448116 KFO80602.1 BC084960 AAH84960.1 AQIA01044234 AQIA01044235 AQIA01044236 AQIA01044237 AQIA01044238 AQIA01044239 AQIA01044240 KL215909 KFV67349.1 ABQF01009901 ABQF01009902 GBYX01190737 JAO82928.1 ADFV01100849 ADFV01100850 ADFV01100851 ADFV01100852 ADFV01100853 AAMC01059311 AAMC01059312 AAMC01059313 AAMC01059314 AAMC01059315 BC136174 AAI36175.1 KK733726 KFQ98318.1 KB031079 ELK03587.1 MUZQ01000222 OWK54982.1 QUSF01000070 RLV96459.1 KE164160 EPQ15582.1 AQIB01146488 AQIB01146489 JSUE03026975 JSUE03026976 AJFE02084028 AJFE02084029 AACZ04066235 GABC01006496 GABF01000977 GABD01004812 GABE01008281 NBAG03000265 JAA04842.1 JAA21168.1 JAA28288.1 JAA36458.1 PNI55893.1 PNI55896.1 PNI55899.1 AANG04004523 NDHI03003657 PNJ11446.1 ABGA01147248 ABGA01147249 ABGA01147250 ABGA01147251 ABGA01147252 PNJ11440.1 PNJ11444.1 HAEH01022424 SBS13938.1 KK719543 KFO64056.1 AAKN02049546 AAKN02049547
OWR48602.1 KQ459580 KPI99250.1 KK520006 KFP66068.1 AADN05000347 AGCU01020983 AGCU01020984 AGCU01020985 KL206176 KFV79635.1 KK534807 KFP29099.1 AZIM01000162 ETE72898.1 AF403117 AF403121 AAL01312.1 AF403115 AF403119 AJ719343 AAL01310.1 CAG31002.1 KL217603 KFO96884.1 KN125754 KFU84322.1 KK501508 KFP15670.1 KL670089 KFW78257.1 KK630484 KFV55417.1 AHZZ02021396 AHZZ02021397 KL410606 KFQ97843.1 KK398741 KFV58723.1 KL895122 KGL82296.1 AKHW03003682 KYO33572.1 KL873342 KGL98378.1 KK438364 KFQ69425.1 GAMS01000760 GAMR01004025 GAMQ01004428 JAB22376.1 JAB29907.1 JAB37423.1 KL226224 KFM10122.1 KL224844 KFW64847.1 KK683546 KFQ13626.1 ADON01053263 GBYX01190734 JAO82930.1 CM004482 OCT63787.1 KL448116 KFO80602.1 BC084960 AAH84960.1 AQIA01044234 AQIA01044235 AQIA01044236 AQIA01044237 AQIA01044238 AQIA01044239 AQIA01044240 KL215909 KFV67349.1 ABQF01009901 ABQF01009902 GBYX01190737 JAO82928.1 ADFV01100849 ADFV01100850 ADFV01100851 ADFV01100852 ADFV01100853 AAMC01059311 AAMC01059312 AAMC01059313 AAMC01059314 AAMC01059315 BC136174 AAI36175.1 KK733726 KFQ98318.1 KB031079 ELK03587.1 MUZQ01000222 OWK54982.1 QUSF01000070 RLV96459.1 KE164160 EPQ15582.1 AQIB01146488 AQIB01146489 JSUE03026975 JSUE03026976 AJFE02084028 AJFE02084029 AACZ04066235 GABC01006496 GABF01000977 GABD01004812 GABE01008281 NBAG03000265 JAA04842.1 JAA21168.1 JAA28288.1 JAA36458.1 PNI55893.1 PNI55896.1 PNI55899.1 AANG04004523 NDHI03003657 PNJ11446.1 ABGA01147248 ABGA01147249 ABGA01147250 ABGA01147251 ABGA01147252 PNJ11440.1 PNJ11444.1 HAEH01022424 SBS13938.1 KK719543 KFO64056.1 AAKN02049546 AAKN02049547
Proteomes
UP000005204
UP000007151
UP000053268
UP000000539
UP000007267
UP000233220
+ More
UP000053584 UP000054308 UP000053119 UP000189705 UP000053258 UP000028761 UP000053283 UP000053641 UP000050525 UP000233020 UP000261360 UP000261420 UP000233040 UP000053858 UP000008225 UP000053286 UP000054081 UP000016666 UP000186698 UP000053760 UP000233100 UP000053875 UP000007754 UP000001646 UP000001073 UP000002279 UP000264820 UP000008143 UP000053605 UP000010552 UP000197619 UP000265120 UP000276834 UP000233120 UP000029965 UP000233060 UP000006718 UP000240080 UP000002277 UP000011712 UP000233200 UP000233080 UP000005226 UP000001595 UP000233180 UP000052976 UP000261620 UP000005447
UP000053584 UP000054308 UP000053119 UP000189705 UP000053258 UP000028761 UP000053283 UP000053641 UP000050525 UP000233020 UP000261360 UP000261420 UP000233040 UP000053858 UP000008225 UP000053286 UP000054081 UP000016666 UP000186698 UP000053760 UP000233100 UP000053875 UP000007754 UP000001646 UP000001073 UP000002279 UP000264820 UP000008143 UP000053605 UP000010552 UP000197619 UP000265120 UP000276834 UP000233120 UP000029965 UP000233060 UP000006718 UP000240080 UP000002277 UP000011712 UP000233200 UP000233080 UP000005226 UP000001595 UP000233180 UP000052976 UP000261620 UP000005447
PRIDE
SUPFAM
SSF50353
SSF50353
CDD
ProteinModelPortal
H9JRN5
A0A212F4F2
A0A194Q779
A0A091M2P3
A0A1D5PS41
K7FFP1
+ More
K7FFR2 A0A2K6SLH3 A0A2K6SLK0 A0A093HJK0 A0A091KMX0 V8PFX0 Q90VV7 Q90VZ5 A0A091HNZ3 A0A093DEP7 A0A091J7G2 A0A3Q0GE77 A0A1U7RXQ4 A0A093Q1Y1 A0A093FCI1 A0A2I3M1R9 A0A091V4A8 A0A093HW59 A0A099ZKD0 A0A151N9R2 A0A2K5CS14 A0A3B4Y9H6 A0A3B4U6H5 A0A2K5R875 A0A2K5CS27 A0A3Q0GEE6 A0A0A0AXY6 A0A091T6F8 U3DFM0 A0A087R9G8 A0A2K5R884 A0A093NXM0 A0A091Q284 U3J7Q1 A0A0S7L0C3 A0A1L8EWQ6 A0A091H2Q2 Q5U4T4 A0A3B4Y9J8 A0A2K5UG64 F7IS82 A0A096NIB1 A0A2Y9SBA4 A0A093GKP7 H0ZLN7 G1KJC4 A0A0S7L091 A0A2I3GLS2 F7FMP3 A0A3Q2Y3Q7 A4IIV9 A0A091V5K9 L5JX02 A0A218UNP1 A0A3P8WSX3 A0A3L8S3D5 S7Q3W8 A0A2K6BU96 A0A2K5UFW6 A0A0D9R561 A0A2K5LEP2 F7G963 A0A2R8ZY51 H2QZM5 A0A2K6BU98 A0A2K5UG24 A0A2I3LDH1 A0A2K5LEX0 A0A1D5QUE4 A0A2R8ZTH9 A0A2I3T8K9 A0A2I2V5I5 A0A2K6RDI1 A0A2K5HB13 G1QQ87 H2SWG8 A0A2K5HAZ8 A0A2J8RSE4 A0A2K6RDM5 A0A2J8RSD5 A0A1A8S8J9 A0A2K6L3P3 A0A091F477 A0A3Q4AR44 H0W0L7 A0A2K6RDG1 A0A3P8WQX3
K7FFR2 A0A2K6SLH3 A0A2K6SLK0 A0A093HJK0 A0A091KMX0 V8PFX0 Q90VV7 Q90VZ5 A0A091HNZ3 A0A093DEP7 A0A091J7G2 A0A3Q0GE77 A0A1U7RXQ4 A0A093Q1Y1 A0A093FCI1 A0A2I3M1R9 A0A091V4A8 A0A093HW59 A0A099ZKD0 A0A151N9R2 A0A2K5CS14 A0A3B4Y9H6 A0A3B4U6H5 A0A2K5R875 A0A2K5CS27 A0A3Q0GEE6 A0A0A0AXY6 A0A091T6F8 U3DFM0 A0A087R9G8 A0A2K5R884 A0A093NXM0 A0A091Q284 U3J7Q1 A0A0S7L0C3 A0A1L8EWQ6 A0A091H2Q2 Q5U4T4 A0A3B4Y9J8 A0A2K5UG64 F7IS82 A0A096NIB1 A0A2Y9SBA4 A0A093GKP7 H0ZLN7 G1KJC4 A0A0S7L091 A0A2I3GLS2 F7FMP3 A0A3Q2Y3Q7 A4IIV9 A0A091V5K9 L5JX02 A0A218UNP1 A0A3P8WSX3 A0A3L8S3D5 S7Q3W8 A0A2K6BU96 A0A2K5UFW6 A0A0D9R561 A0A2K5LEP2 F7G963 A0A2R8ZY51 H2QZM5 A0A2K6BU98 A0A2K5UG24 A0A2I3LDH1 A0A2K5LEX0 A0A1D5QUE4 A0A2R8ZTH9 A0A2I3T8K9 A0A2I2V5I5 A0A2K6RDI1 A0A2K5HB13 G1QQ87 H2SWG8 A0A2K5HAZ8 A0A2J8RSE4 A0A2K6RDM5 A0A2J8RSD5 A0A1A8S8J9 A0A2K6L3P3 A0A091F477 A0A3Q4AR44 H0W0L7 A0A2K6RDG1 A0A3P8WQX3
PDB
5HP3
E-value=1.09035e-80,
Score=766
Ontologies
GO
GO:0004000
GO:0006396
GO:0003723
GO:0060384
GO:0006382
GO:0097049
GO:0005654
GO:0051726
GO:0061744
GO:0007274
GO:0045070
GO:0060415
GO:0008285
GO:0005829
GO:0021965
GO:0005730
GO:0021618
GO:0050884
GO:0030336
GO:0021610
GO:0035264
GO:0044387
GO:0003726
GO:0003725
GO:0005634
GO:0005737
GO:0008251
GO:0006508
GO:0016701
GO:0006355
GO:0003824
GO:0008152
GO:0016491
GO:0015930
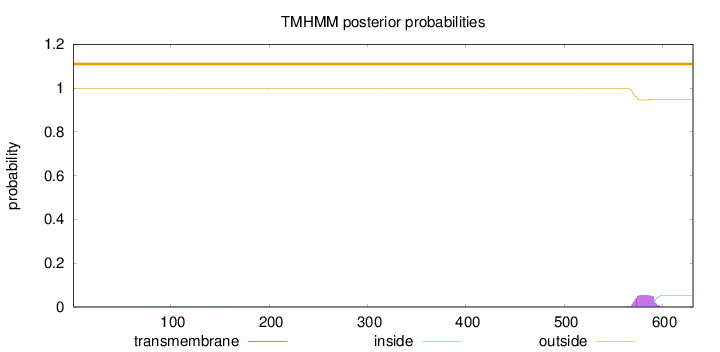
Topology
Length:
631
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.19675
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00314
outside
1 - 631
Population Genetic Test Statistics
Pi
167.210517
Theta
143.851008
Tajima's D
0.290684
CLR
12.572953
CSRT
0.448277586120694
Interpretation
Uncertain
