Gene
KWMTBOMO14842
Annotation
PREDICTED:_pancreatic_lipase-related_protein_2-like_[Papilio_polytes]
Location in the cell
PlasmaMembrane Reliability : 1.873
Sequence
CDS
ATGCAGACAGCAATTCCTTTGGCGTGTCTTTTATCCGTCATAAGACCACCAGGAGTGCAGTTCGAGAACTCTTTGCTGCAAATAGCGCCCATCGACAAGTCAAAGTGCCCCTTCGTGAAGAACACCAATGATGTTGGTTTCCAGCTATACACTAGGCATAACCCGACAGTCTACCAGGAGCTGGTCTACGGTGATGACGAGAAGCTCTTTGCTTCGAATATAGACTTCAACGATAAGACGGTTCTCTACTTCCACGCGTTCATGGAGCAGCCCGACGACGGTAGCGGCATCATGATCAGGGAAGCATACGTCCAGCGAGGTGACACGAACGTCATCATGATAGACGCTCATCATCTCGAAGCGGGACCGTGGTACGTCACGGCGGCCCAGAACACGTGGTACATAGGACGCTTCGCGGCGCAGTTCATCGACTTCCTCGTGACCAGGGGGCTGAATCTAAACAGCACCCATCTGATAGGGCACAGCCTCGGGGCTCAGAGCGCCGGTGTCGCCGGTAGCTCATTGAAATCAGGTCGCGTGGCCAGAATAACGGGCCTAGATCCGGCGCTGCCGTTATTCGAAGGCCTACCGATAGACCAGCGTCTGGACGCATCGGACGCCGAATTCGTTGACGTTATTCACACCGACGCCGGTATTTTTGGATACAAGGCGCCGATCGGTCACGTGGACTTTTACCCGAACGGCGGCATCAGTCCCCAGCCAGGATGCGAGCTGGAAGCCGTCATACCACAGCAGCTACTTCTGAACAAGTTCTTCTGCAGCCACTGGCGTTCATATCAGTTCTACACGGAATCTGTAATCCGGCCGCGTTCATTCCTCGCCACCGAATGCACCTCGTGGAGGCGGTACTCCAGTGGTGGTTGCGAATTCGAGAAAACCACACACATGGGATTCGGGGTCGATACGAAGGTTTACGAATGTATAGTTATATATGACCACATGTCTATTGTCATGACAACAAATTTACTATACTTATAA
Protein
MQTAIPLACLLSVIRPPGVQFENSLLQIAPIDKSKCPFVKNTNDVGFQLYTRHNPTVYQELVYGDDEKLFASNIDFNDKTVLYFHAFMEQPDDGSGIMIREAYVQRGDTNVIMIDAHHLEAGPWYVTAAQNTWYIGRFAAQFIDFLVTRGLNLNSTHLIGHSLGAQSAGVAGSSLKSGRVARITGLDPALPLFEGLPIDQRLDASDAEFVDVIHTDAGIFGYKAPIGHVDFYPNGGISPQPGCELEAVIPQQLLLNKFFCSHWRSYQFYTESVIRPRSFLATECTSWRRYSSGGCEFEKTTHMGFGVDTKVYECIVIYDHMSIVMTTNLLYL
Summary
Similarity
Belongs to the AB hydrolase superfamily. Lipase family.
Uniprot
A0A0N1PJG6
A0A194Q121
A0A2W1B8G3
A0A2A4J568
D6WBV6
A0A1W4X7R5
+ More
U4UUN0 A0A2J7QJ67 A0A067QPW6 A0A2S2QRV2 A0A2S2PCJ6 A0A0A9XTP0 A0A1B0EZU1 A0A146LFH2 A0A1I8PID3 A0A154P1R9 A0A182N277 A0A1I8PIC9 A0A084WM82 A0A182JG03 N6T1A6 A0A088AM94 Q17PN5 A0A1B0BVR0 A0A182FC01 A0A2H8TRV5 A0A1I8MK50 A0A182YAZ1 B4JVL5 A0A0J9REI3 A0A182GSZ6 B4QIH5 A0A182QNU7 A0A182PJM6 J9JWA0 A0A1I8MK53 A0A1A9ZP98 W5J3D4 A0A1A9XLC0 B4HTB1 A0A182RAL2 A0A151I0K8 A1ZAL5 A0A1W4UZL6 A0A3L8DSC5 B3MD95 A0A182KKY7 B3NP53 A0A182VGE1 A0A0Q5VLT7 B4LP71 A0A3B0J1K3 A0A182MU85 A0A182WDM4 Q7QHC0 A0A1B0FLI3 A0A0R1DYC0 B4P5U1 A0A182HW84 A0A1I8PIH6 A0A0R3NTI7 A0A195DPR4 A0A1A9VX61 A0A232EVI5 A0A1A9WK45 B4MR93 A0A1J1I7R1 A0A151X5R7 B4KSQ9 A0A0M9A1K6 A0A0A1XE33 A0A0L0CMA1 A0A0Q9W2T7 A0A182UIZ6 A0A154PEY0 A0A1B6C5E1 A0A194Q8Y1 A0A1B6DU95 A0A212EP55 A0A034WE39 E1ZYV2 A0A0M3QVQ4 A0A0M4EKW3 A0A0L7QY80 A0A3B0JUT0 A0A2A3EET9 K7IWE9 N6TY43 A0A2A4JW95 A0A0K8VE94 A0A087ZYV7 A0A2W1BTQ3 E2BMN0 E0VE21 W8C6D4 A0A1Y1KB23
U4UUN0 A0A2J7QJ67 A0A067QPW6 A0A2S2QRV2 A0A2S2PCJ6 A0A0A9XTP0 A0A1B0EZU1 A0A146LFH2 A0A1I8PID3 A0A154P1R9 A0A182N277 A0A1I8PIC9 A0A084WM82 A0A182JG03 N6T1A6 A0A088AM94 Q17PN5 A0A1B0BVR0 A0A182FC01 A0A2H8TRV5 A0A1I8MK50 A0A182YAZ1 B4JVL5 A0A0J9REI3 A0A182GSZ6 B4QIH5 A0A182QNU7 A0A182PJM6 J9JWA0 A0A1I8MK53 A0A1A9ZP98 W5J3D4 A0A1A9XLC0 B4HTB1 A0A182RAL2 A0A151I0K8 A1ZAL5 A0A1W4UZL6 A0A3L8DSC5 B3MD95 A0A182KKY7 B3NP53 A0A182VGE1 A0A0Q5VLT7 B4LP71 A0A3B0J1K3 A0A182MU85 A0A182WDM4 Q7QHC0 A0A1B0FLI3 A0A0R1DYC0 B4P5U1 A0A182HW84 A0A1I8PIH6 A0A0R3NTI7 A0A195DPR4 A0A1A9VX61 A0A232EVI5 A0A1A9WK45 B4MR93 A0A1J1I7R1 A0A151X5R7 B4KSQ9 A0A0M9A1K6 A0A0A1XE33 A0A0L0CMA1 A0A0Q9W2T7 A0A182UIZ6 A0A154PEY0 A0A1B6C5E1 A0A194Q8Y1 A0A1B6DU95 A0A212EP55 A0A034WE39 E1ZYV2 A0A0M3QVQ4 A0A0M4EKW3 A0A0L7QY80 A0A3B0JUT0 A0A2A3EET9 K7IWE9 N6TY43 A0A2A4JW95 A0A0K8VE94 A0A087ZYV7 A0A2W1BTQ3 E2BMN0 E0VE21 W8C6D4 A0A1Y1KB23
Pubmed
26354079
28756777
18362917
19820115
23537049
24845553
+ More
25401762 26823975 24438588 17510324 25315136 25244985 17994087 22936249 26483478 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 30249741 20966253 12364791 14747013 17210077 17550304 15632085 28648823 25830018 26108605 22118469 25348373 20798317 20075255 20566863 24495485 28004739
25401762 26823975 24438588 17510324 25315136 25244985 17994087 22936249 26483478 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 30249741 20966253 12364791 14747013 17210077 17550304 15632085 28648823 25830018 26108605 22118469 25348373 20798317 20075255 20566863 24495485 28004739
EMBL
KQ460240
KPJ16392.1
KQ459580
KPI99251.1
KZ150567
PZC70514.1
+ More
NWSH01003138 PCG66886.1 KQ971315 EEZ97864.2 KB632375 ERL93896.1 NEVH01013559 PNF28635.1 KK853686 KDQ97822.1 GGMS01010649 MBY79852.1 GGMR01014558 MBY27177.1 GBHO01020355 JAG23249.1 AJVK01007790 AJVK01007791 GDHC01012863 JAQ05766.1 KQ434796 KZC05783.1 ATLV01024391 KE525351 KFB51326.1 APGK01048183 APGK01048184 APGK01048185 APGK01048186 KB741101 ENN73869.1 CH477190 EAT48716.1 JXJN01021437 GFXV01004143 MBW15948.1 CH916375 EDV98483.1 CM002911 KMY94392.1 JXUM01002342 JXUM01002343 JXUM01002344 JXUM01002345 JXUM01002346 KQ560137 KXJ84231.1 CM000362 EDX07422.1 AXCN02001739 ABLF02035114 ABLF02035118 ADMH02002203 ETN57833.1 CH480816 EDW48212.1 KQ976619 KYM79087.1 AE013599 AAF57935.2 QOIP01000004 RLU23345.1 CH902619 EDV37428.2 CH954179 EDV55692.1 KQS62407.1 CH940648 EDW60180.1 OUUW01000001 SPP74827.1 AXCM01016020 AAAB01008816 EAA05324.4 CCAG010023317 CM000158 KRK00148.1 EDW91857.2 APCN01001475 CM000071 KRT02099.1 KQ980653 KYN14802.1 NNAY01001993 OXU22363.1 CH963849 EDW74632.1 CVRI01000042 CRK95612.1 KQ982494 KYQ55731.1 CH933808 EDW10558.2 KQ435765 KOX75317.1 GBXI01007275 GBXI01005070 GBXI01004777 JAD07017.1 JAD09222.1 JAD09515.1 JRES01000195 KNC33420.1 KRF79293.1 KQ434875 KZC09780.1 GEDC01028789 JAS08509.1 KQ459562 KPI99875.1 GEDC01008089 JAS29209.1 AGBW02013528 OWR43264.1 GAKP01006370 JAC52582.1 GL435242 EFN73642.1 CP012524 ALC42809.1 ALC42105.1 KQ414697 KOC63514.1 SPP74828.1 KZ288269 PBC29994.1 APGK01056924 APGK01056925 KB741277 KB632155 ENN71207.1 ERL89184.1 NWSH01000489 PCG76056.1 GDHF01015142 GDHF01012905 JAI37172.1 JAI39409.1 KZ149902 PZC78448.1 GL449275 EFN83048.1 DS235088 EEB11627.1 GAMC01004329 JAC02227.1 GEZM01092065 JAV56666.1
NWSH01003138 PCG66886.1 KQ971315 EEZ97864.2 KB632375 ERL93896.1 NEVH01013559 PNF28635.1 KK853686 KDQ97822.1 GGMS01010649 MBY79852.1 GGMR01014558 MBY27177.1 GBHO01020355 JAG23249.1 AJVK01007790 AJVK01007791 GDHC01012863 JAQ05766.1 KQ434796 KZC05783.1 ATLV01024391 KE525351 KFB51326.1 APGK01048183 APGK01048184 APGK01048185 APGK01048186 KB741101 ENN73869.1 CH477190 EAT48716.1 JXJN01021437 GFXV01004143 MBW15948.1 CH916375 EDV98483.1 CM002911 KMY94392.1 JXUM01002342 JXUM01002343 JXUM01002344 JXUM01002345 JXUM01002346 KQ560137 KXJ84231.1 CM000362 EDX07422.1 AXCN02001739 ABLF02035114 ABLF02035118 ADMH02002203 ETN57833.1 CH480816 EDW48212.1 KQ976619 KYM79087.1 AE013599 AAF57935.2 QOIP01000004 RLU23345.1 CH902619 EDV37428.2 CH954179 EDV55692.1 KQS62407.1 CH940648 EDW60180.1 OUUW01000001 SPP74827.1 AXCM01016020 AAAB01008816 EAA05324.4 CCAG010023317 CM000158 KRK00148.1 EDW91857.2 APCN01001475 CM000071 KRT02099.1 KQ980653 KYN14802.1 NNAY01001993 OXU22363.1 CH963849 EDW74632.1 CVRI01000042 CRK95612.1 KQ982494 KYQ55731.1 CH933808 EDW10558.2 KQ435765 KOX75317.1 GBXI01007275 GBXI01005070 GBXI01004777 JAD07017.1 JAD09222.1 JAD09515.1 JRES01000195 KNC33420.1 KRF79293.1 KQ434875 KZC09780.1 GEDC01028789 JAS08509.1 KQ459562 KPI99875.1 GEDC01008089 JAS29209.1 AGBW02013528 OWR43264.1 GAKP01006370 JAC52582.1 GL435242 EFN73642.1 CP012524 ALC42809.1 ALC42105.1 KQ414697 KOC63514.1 SPP74828.1 KZ288269 PBC29994.1 APGK01056924 APGK01056925 KB741277 KB632155 ENN71207.1 ERL89184.1 NWSH01000489 PCG76056.1 GDHF01015142 GDHF01012905 JAI37172.1 JAI39409.1 KZ149902 PZC78448.1 GL449275 EFN83048.1 DS235088 EEB11627.1 GAMC01004329 JAC02227.1 GEZM01092065 JAV56666.1
Proteomes
UP000053240
UP000053268
UP000218220
UP000007266
UP000192223
UP000030742
+ More
UP000235965 UP000027135 UP000092462 UP000095300 UP000076502 UP000075884 UP000030765 UP000075880 UP000019118 UP000005203 UP000008820 UP000092460 UP000069272 UP000095301 UP000076408 UP000001070 UP000069940 UP000249989 UP000000304 UP000075886 UP000075885 UP000007819 UP000092445 UP000000673 UP000092443 UP000001292 UP000075900 UP000078540 UP000000803 UP000192221 UP000279307 UP000007801 UP000075882 UP000008711 UP000075903 UP000008792 UP000268350 UP000075883 UP000075920 UP000007062 UP000092444 UP000002282 UP000075840 UP000001819 UP000078492 UP000078200 UP000215335 UP000091820 UP000007798 UP000183832 UP000075809 UP000009192 UP000053105 UP000037069 UP000075902 UP000007151 UP000000311 UP000092553 UP000053825 UP000242457 UP000002358 UP000008237 UP000009046
UP000235965 UP000027135 UP000092462 UP000095300 UP000076502 UP000075884 UP000030765 UP000075880 UP000019118 UP000005203 UP000008820 UP000092460 UP000069272 UP000095301 UP000076408 UP000001070 UP000069940 UP000249989 UP000000304 UP000075886 UP000075885 UP000007819 UP000092445 UP000000673 UP000092443 UP000001292 UP000075900 UP000078540 UP000000803 UP000192221 UP000279307 UP000007801 UP000075882 UP000008711 UP000075903 UP000008792 UP000268350 UP000075883 UP000075920 UP000007062 UP000092444 UP000002282 UP000075840 UP000001819 UP000078492 UP000078200 UP000215335 UP000091820 UP000007798 UP000183832 UP000075809 UP000009192 UP000053105 UP000037069 UP000075902 UP000007151 UP000000311 UP000092553 UP000053825 UP000242457 UP000002358 UP000008237 UP000009046
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A0N1PJG6
A0A194Q121
A0A2W1B8G3
A0A2A4J568
D6WBV6
A0A1W4X7R5
+ More
U4UUN0 A0A2J7QJ67 A0A067QPW6 A0A2S2QRV2 A0A2S2PCJ6 A0A0A9XTP0 A0A1B0EZU1 A0A146LFH2 A0A1I8PID3 A0A154P1R9 A0A182N277 A0A1I8PIC9 A0A084WM82 A0A182JG03 N6T1A6 A0A088AM94 Q17PN5 A0A1B0BVR0 A0A182FC01 A0A2H8TRV5 A0A1I8MK50 A0A182YAZ1 B4JVL5 A0A0J9REI3 A0A182GSZ6 B4QIH5 A0A182QNU7 A0A182PJM6 J9JWA0 A0A1I8MK53 A0A1A9ZP98 W5J3D4 A0A1A9XLC0 B4HTB1 A0A182RAL2 A0A151I0K8 A1ZAL5 A0A1W4UZL6 A0A3L8DSC5 B3MD95 A0A182KKY7 B3NP53 A0A182VGE1 A0A0Q5VLT7 B4LP71 A0A3B0J1K3 A0A182MU85 A0A182WDM4 Q7QHC0 A0A1B0FLI3 A0A0R1DYC0 B4P5U1 A0A182HW84 A0A1I8PIH6 A0A0R3NTI7 A0A195DPR4 A0A1A9VX61 A0A232EVI5 A0A1A9WK45 B4MR93 A0A1J1I7R1 A0A151X5R7 B4KSQ9 A0A0M9A1K6 A0A0A1XE33 A0A0L0CMA1 A0A0Q9W2T7 A0A182UIZ6 A0A154PEY0 A0A1B6C5E1 A0A194Q8Y1 A0A1B6DU95 A0A212EP55 A0A034WE39 E1ZYV2 A0A0M3QVQ4 A0A0M4EKW3 A0A0L7QY80 A0A3B0JUT0 A0A2A3EET9 K7IWE9 N6TY43 A0A2A4JW95 A0A0K8VE94 A0A087ZYV7 A0A2W1BTQ3 E2BMN0 E0VE21 W8C6D4 A0A1Y1KB23
U4UUN0 A0A2J7QJ67 A0A067QPW6 A0A2S2QRV2 A0A2S2PCJ6 A0A0A9XTP0 A0A1B0EZU1 A0A146LFH2 A0A1I8PID3 A0A154P1R9 A0A182N277 A0A1I8PIC9 A0A084WM82 A0A182JG03 N6T1A6 A0A088AM94 Q17PN5 A0A1B0BVR0 A0A182FC01 A0A2H8TRV5 A0A1I8MK50 A0A182YAZ1 B4JVL5 A0A0J9REI3 A0A182GSZ6 B4QIH5 A0A182QNU7 A0A182PJM6 J9JWA0 A0A1I8MK53 A0A1A9ZP98 W5J3D4 A0A1A9XLC0 B4HTB1 A0A182RAL2 A0A151I0K8 A1ZAL5 A0A1W4UZL6 A0A3L8DSC5 B3MD95 A0A182KKY7 B3NP53 A0A182VGE1 A0A0Q5VLT7 B4LP71 A0A3B0J1K3 A0A182MU85 A0A182WDM4 Q7QHC0 A0A1B0FLI3 A0A0R1DYC0 B4P5U1 A0A182HW84 A0A1I8PIH6 A0A0R3NTI7 A0A195DPR4 A0A1A9VX61 A0A232EVI5 A0A1A9WK45 B4MR93 A0A1J1I7R1 A0A151X5R7 B4KSQ9 A0A0M9A1K6 A0A0A1XE33 A0A0L0CMA1 A0A0Q9W2T7 A0A182UIZ6 A0A154PEY0 A0A1B6C5E1 A0A194Q8Y1 A0A1B6DU95 A0A212EP55 A0A034WE39 E1ZYV2 A0A0M3QVQ4 A0A0M4EKW3 A0A0L7QY80 A0A3B0JUT0 A0A2A3EET9 K7IWE9 N6TY43 A0A2A4JW95 A0A0K8VE94 A0A087ZYV7 A0A2W1BTQ3 E2BMN0 E0VE21 W8C6D4 A0A1Y1KB23
PDB
1BU8
E-value=2.44667e-41,
Score=424
Ontologies
GO
PANTHER
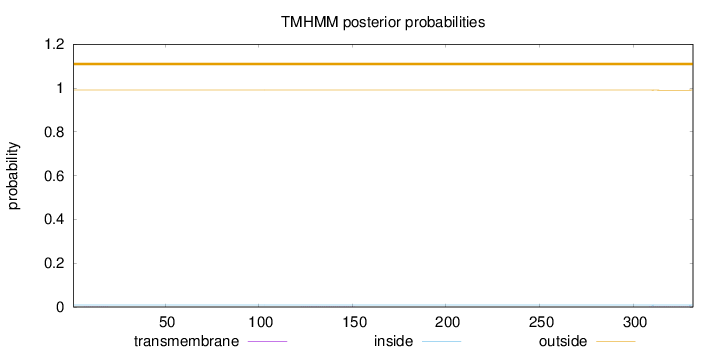
Topology
Subcellular location
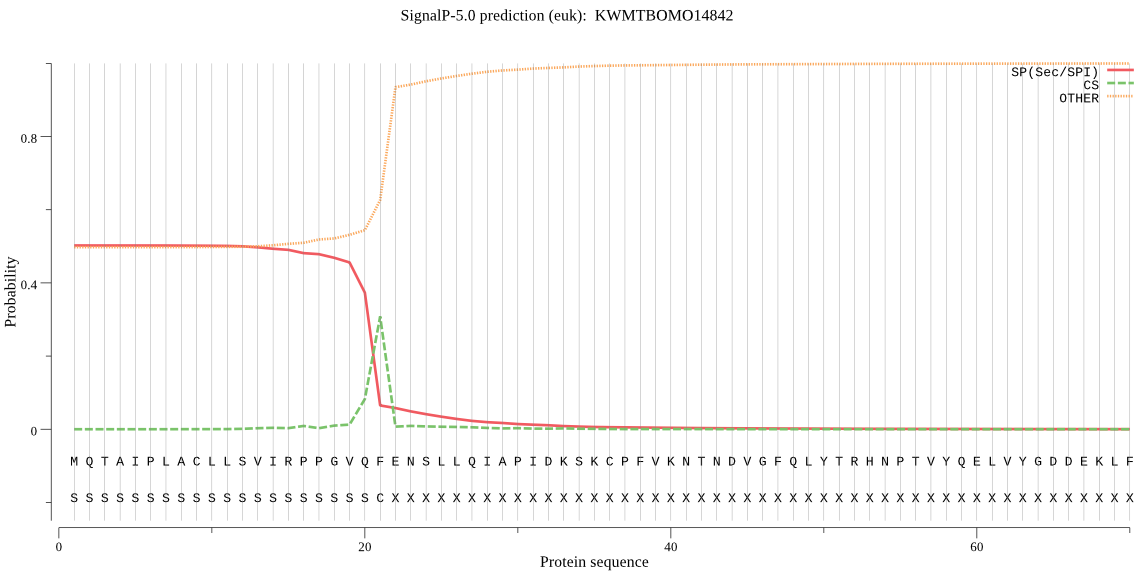
Secreted
SignalP
Position: 1 - 21,
Likelihood: 0.502177
Length:
332
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02779
Exp number, first 60 AAs:
0.00267
Total prob of N-in:
0.00911
outside
1 - 332
Population Genetic Test Statistics
Pi
231.672223
Theta
185.183123
Tajima's D
1.152659
CLR
0.609334
CSRT
0.698765061746913
Interpretation
Uncertain
