Pre Gene Modal
BGIBMGA012145
Annotation
PREDICTED:_probable_tyrosyl-DNA_phosphodiesterase_[Amyelois_transitella]
Full name
Mitochondrial genome maintenance exonuclease 1
+ More
Probable tyrosyl-DNA phosphodiesterase
Probable tyrosyl-DNA phosphodiesterase
Alternative Name
Protein glaikit
Location in the cell
Nuclear Reliability : 2.803
Sequence
CDS
ATGAACCCTTCCCTAAAGCGACGAGGAACTATGCAAGAAAATGAGACAAAAAGAGTAAAAAAGGTTTGTGATTATGGCGAAAAATGCTACAGAATGAATCCTGTACATTTCCGTGAATTTAGTCATCCGCATTTGGAATCTATATTGGATAACCATGCCAGTGGTGGAGATTACCCTATCCCAGATAAATACAATCTCCAGAAGAAATTGATCACTGAACAGCTAGATCTTATAATCGAAAAGGGATTCTATGCTCCTCGGAACAATGTACAAAACAACCCAAAACAAATTGAAAACAAACAAGAAAGAGACAGCGACAGACGAGTAGGGAAAAATGTGGAACCGGAGGCAAGTTCTGCTAGTCATCAAAAAGTACCGGACAAACCTGAAAATACAAAAACAATGTTTTCAAATAATGAAGCCAAAATTAAGACACCTGTTGATAGAGGGGGAGTTGTTAAAGAGAAATCCACTAACTCAGATTATCGTCCCATAATACCTCCGACTCGACGTGTAGAAGATTATTTGAACGTTGTAAGACCTAAAGGGCGAATGGCTGCAAAGCACGAAGCCAGTGCGCCCTTTTATATTTTTTACACAACAATCACGGCGGCAAAGGAAACTCATTCGCAGCCGTTCTCTATAACATTCCAAGAAATTTTAGATAGAAGCTTAGGCGAGTTGAAGTGTTCGTTGCAAATCAATTTTATGGTCGAACTTGGATGGCTCTTGGCTCAGTACTACTTCGCTGGTTACAGCGAGAAGAAGCTAACGATATTGTACGGTGAGGATTCTCAGGATTTAAGGACGATCAGCCAGAAGAAGCCACACGTTGACGCTCACCTGGTGCCGATGGCGACTCCGTTCGGTAAACATCACACTAAAATGATGATCTTGTGTTACGAAGATGGGTCACTTCGCGTTGTAGTATCGACAGCCAATCTGTATATTGATGACTGGGAGAACCGAACGCAAGGTCTATGGTTCAGTCCAAAATGCCCAGAGTTGCCGCCGGAAGCGATGCCTCATGATGGAGAGTCTCCTACGATGTTCAAAAAGTCTCTGTTACGCTATTTGAATCACTATCATATGCCTTATCTCACCTACTACGTTGAAAGAGTGAAGAGATCGGATTTCAGTCATATTAATGTTTTCTTAGTAGCATCGGCTCCTGGATCTCATTTTGACATGGACTGGGGCATGACCCGTGTGGGCTCATTACTGCGGCAGCACTGCTGTATACCACCCGAGGAACAGCTCCAGTGGCCCCTGGTGGCACAGGCCAGCAGCCTGGGTAGTTACGGCAAGGATCCGAAGCTCTGGTTGACCGGAGATTTCCTTCACAACTTCACGAAAATCAAGAACCAATCGCAGATGTTGTCGTCTCCGCCTACTTTGAAACTGATTTATCCATCTTTAGAAAACGTCAAACAATCTCACGACGATTTGTTAGGAGGCGGTTGCTTGCCTTACGCAGCCGAAGCGCATTCGAAACAACCTTGGCTAAATAGCTTTTTATATCAATGGCGAGCTGCGTCCACGAATCGAAACCGGGCGATGCCCCACATAAAGTCCTACACCCGGGTGTCGAAGGACGGCAGGAAGGCAGCGTACTACCTGCTCACGTCGGGGAACGTCAGCAAGGCGGCCTGGGGCTCGACGAACAAAGGAAACGGAGCGCTCAGGATAATGAGTTACGAAGCCGGCGTGCTGTTCCTGCCAAAGTTCATTACCAACGAAGATTACTTCTGTCTAGAACAAAACGCCCCGAACCGGCTTATCGTGCCATACGACTTACCGCCAGTCAAATATACGGATGGCATGTCGCCCTGGGTCTCTGATTACCTCATGTGA
Protein
MNPSLKRRGTMQENETKRVKKVCDYGEKCYRMNPVHFREFSHPHLESILDNHASGGDYPIPDKYNLQKKLITEQLDLIIEKGFYAPRNNVQNNPKQIENKQERDSDRRVGKNVEPEASSASHQKVPDKPENTKTMFSNNEAKIKTPVDRGGVVKEKSTNSDYRPIIPPTRRVEDYLNVVRPKGRMAAKHEASAPFYIFYTTITAAKETHSQPFSITFQEILDRSLGELKCSLQINFMVELGWLLAQYYFAGYSEKKLTILYGEDSQDLRTISQKKPHVDAHLVPMATPFGKHHTKMMILCYEDGSLRVVVSTANLYIDDWENRTQGLWFSPKCPELPPEAMPHDGESPTMFKKSLLRYLNHYHMPYLTYYVERVKRSDFSHINVFLVASAPGSHFDMDWGMTRVGSLLRQHCCIPPEEQLQWPLVAQASSLGSYGKDPKLWLTGDFLHNFTKIKNQSQMLSSPPTLKLIYPSLENVKQSHDDLLGGGCLPYAAEAHSKQPWLNSFLYQWRAASTNRNRAMPHIKSYTRVSKDGRKAAYYLLTSGNVSKAAWGSTNKGNGALRIMSYEAGVLFLPKFITNEDYFCLEQNAPNRLIVPYDLPPVKYTDGMSPWVSDYLM
Summary
Description
Metal-dependent single-stranded DNA (ssDNA) exonuclease involved in mitochondrial genome maintenance.
DNA repair enzyme that can remove a variety of covalent adducts from DNA through hydrolysis of a 3'-phosphodiester bond, giving rise to DNA with a free 3' phosphate. Catalyzes the hydrolysis of dead-end complexes between DNA and the topoisomerase I active site tyrosine residue. Hydrolyzes 3'-phosphoglycolates on protruding 3' ends on DNA double-strand breaks due to DNA damage by radiation and free radicals. Acts on blunt-ended double-strand DNA breaks and on single-stranded DNA. May have low 3'exonuclease activity and may be able to remove a single nucleoside from the 3'end of DNA and RNA molecules with 3'hydroxyl groups. Has no exonuclease activity towards DNA or RNA with a 3'phosphate (By similarity). Required for normal polarization of epidermal cells, correct subcellular location of the Crb complex to the apical lateral membrane, and for normal neuronal development during embryonic development.
DNA repair enzyme that can remove a variety of covalent adducts from DNA through hydrolysis of a 3'-phosphodiester bond, giving rise to DNA with a free 3' phosphate. Catalyzes the hydrolysis of dead-end complexes between DNA and the topoisomerase I active site tyrosine residue. Hydrolyzes 3'-phosphoglycolates on protruding 3' ends on DNA double-strand breaks due to DNA damage by radiation and free radicals. Acts on blunt-ended double-strand DNA breaks and on single-stranded DNA. May have low 3'exonuclease activity and may be able to remove a single nucleoside from the 3'end of DNA and RNA molecules with 3'hydroxyl groups. Has no exonuclease activity towards DNA or RNA with a 3'phosphate (By similarity). Required for normal polarization of epidermal cells, correct subcellular location of the Crb complex to the apical lateral membrane, and for normal neuronal development during embryonic development.
Similarity
Belongs to the MGME1 family.
Belongs to the tyrosyl-DNA phosphodiesterase family.
Belongs to the tyrosyl-DNA phosphodiesterase family.
Keywords
Complete proteome
Cytoplasm
DNA damage
DNA repair
Exonuclease
Hydrolase
Nuclease
Nucleus
Reference proteome
Repeat
Feature
chain Probable tyrosyl-DNA phosphodiesterase
Uniprot
H9JRI5
A0A0N0PDC5
A0A212FLB5
S4NP13
A0A2W1BI46
A0A1B0DJS2
+ More
Q5TSF5 A0A1Y9GLD9 A0A182G8A8 B4KEU9 A0A182VGJ1 W5JHK8 A0A182S510 A0A182YA38 B4Q941 A0A1Q3EYJ2 A0A1Q3EYJ3 B4MWZ6 A0A182LQ33 A0A182QUU8 A0A3B0KF10 A0A084WI56 B4NXE5 A0A1I8PBS7 B3NAR4 Q9VQM4 A0A182ITF6 B3N203 Q29L30 B4JAN6 A0A182X1U4 A0A0M4EP57 B4LU32 A0A182FV04 A0A1W4V7I2 A0A182TNC9 A0A182JRK8 A0A1I8N345 A0A182P8G8 A0A336LXP3 A0A0L0CE48 A0A1B0A4S1 A0A182VTC3 A0A1A9W6E8 A0A1J1J5J5 A0A1B6IMP2 A0A0A1XHB1 A0A2M4APZ2 A0A2M4APT8 A0A2M4APZ9 A0A182SYC7 A0A0K8VE29 A0A1B6ES60 A0A1A9YKG6 A0A034WEA8 Q16PH5 Q16EJ4 A0A1B0FC16 A0A146MFD4 A0A0A9XGY5 A0A182MYE1 B0WGW2 A0A0K8SQK4 A0A2J7PZ96 A0A1I8N346 A0A067R8H0 A0A1B0BPX4 A0A1A9USD2 W8BSM7 A0A1B6HXL7 E9J172 F4W9G0 A0A023F0N4 A0A154NXS3 A0A146MAD7 E1ZZ54 A0A2M4ASB2 A0A0V0GAW7 A0A1Y9GKN2 A0A158N984 A0A026W3D2 T1H9D3 B4I2W4 A0A1S4HEZ1 A0A0L7RKH2 A0A088AUJ5 A0A0P5FIR5 A0A158N982 A0A0M8ZWA3 A0A0P6ACI7 A0A195C978 A0A2A3ESN6 E2B964 A0A1W4XJL3
Q5TSF5 A0A1Y9GLD9 A0A182G8A8 B4KEU9 A0A182VGJ1 W5JHK8 A0A182S510 A0A182YA38 B4Q941 A0A1Q3EYJ2 A0A1Q3EYJ3 B4MWZ6 A0A182LQ33 A0A182QUU8 A0A3B0KF10 A0A084WI56 B4NXE5 A0A1I8PBS7 B3NAR4 Q9VQM4 A0A182ITF6 B3N203 Q29L30 B4JAN6 A0A182X1U4 A0A0M4EP57 B4LU32 A0A182FV04 A0A1W4V7I2 A0A182TNC9 A0A182JRK8 A0A1I8N345 A0A182P8G8 A0A336LXP3 A0A0L0CE48 A0A1B0A4S1 A0A182VTC3 A0A1A9W6E8 A0A1J1J5J5 A0A1B6IMP2 A0A0A1XHB1 A0A2M4APZ2 A0A2M4APT8 A0A2M4APZ9 A0A182SYC7 A0A0K8VE29 A0A1B6ES60 A0A1A9YKG6 A0A034WEA8 Q16PH5 Q16EJ4 A0A1B0FC16 A0A146MFD4 A0A0A9XGY5 A0A182MYE1 B0WGW2 A0A0K8SQK4 A0A2J7PZ96 A0A1I8N346 A0A067R8H0 A0A1B0BPX4 A0A1A9USD2 W8BSM7 A0A1B6HXL7 E9J172 F4W9G0 A0A023F0N4 A0A154NXS3 A0A146MAD7 E1ZZ54 A0A2M4ASB2 A0A0V0GAW7 A0A1Y9GKN2 A0A158N984 A0A026W3D2 T1H9D3 B4I2W4 A0A1S4HEZ1 A0A0L7RKH2 A0A088AUJ5 A0A0P5FIR5 A0A158N982 A0A0M8ZWA3 A0A0P6ACI7 A0A195C978 A0A2A3ESN6 E2B964 A0A1W4XJL3
EC Number
3.1.-.-
3.1.4.-
3.1.4.-
Pubmed
19121390
26354079
22118469
23622113
28756777
12364791
+ More
26483478 17994087 20920257 23761445 25244985 22936249 20966253 24438588 17550304 10940635 10731132 12537572 12537569 15556867 15632085 25315136 26108605 25830018 25348373 17510324 26823975 25401762 24845553 24495485 21282665 21719571 25474469 20798317 21347285 24508170 30249741
26483478 17994087 20920257 23761445 25244985 22936249 20966253 24438588 17550304 10940635 10731132 12537572 12537569 15556867 15632085 25315136 26108605 25830018 25348373 17510324 26823975 25401762 24845553 24495485 21282665 21719571 25474469 20798317 21347285 24508170 30249741
EMBL
BABH01031850
BABH01031851
BABH01031852
BABH01031853
BABH01031854
KQ460396
+ More
KPJ15335.1 AGBW02007808 OWR54526.1 GAIX01012039 JAA80521.1 KZ150076 PZC73951.1 AJVK01001043 AAAB01008933 EAL40355.3 APCN01005565 JXUM01151776 KQ571296 KXJ68209.1 CH933807 EDW12999.1 ADMH02001479 ETN62390.1 CM000361 CM002910 EDX03580.1 KMY87828.1 GFDL01014674 JAV20371.1 GFDL01014671 JAV20374.1 CH963857 EDW76635.1 AXCN02001048 OUUW01000006 SPP82198.1 ATLV01023914 KE525347 KFB49900.1 CM000157 EDW87502.1 CH954177 EDV57587.1 AJ277122 AE014134 AY051884 AY060810 CH902667 EDV30613.1 CH379061 EAL32995.1 CH916368 EDW03844.1 CP012523 ALC38575.1 CH940649 EDW65085.1 UFQT01000279 SSX22705.1 JRES01000507 KNC30525.1 CVRI01000070 CRL07226.1 GECU01019521 JAS88185.1 GBXI01004002 JAD10290.1 GGFK01009544 MBW42865.1 GGFK01009485 MBW42806.1 GGFK01009545 MBW42866.1 GDHF01015186 JAI37128.1 GECZ01029008 JAS40761.1 GAKP01005928 JAC53024.1 CH477781 EAT36273.1 CH478692 EAT32652.1 CCAG010017571 GDHC01001319 JAQ17310.1 GBHO01025551 JAG18053.1 DS231930 EDS27256.1 GBRD01010415 JAG55409.1 NEVH01020337 PNF21657.1 KK852631 KDR19846.1 JXJN01018248 JXJN01023093 GAMC01010272 GAMC01010269 GAMC01010266 JAB96286.1 GECU01028321 JAS79385.1 GL767586 EFZ13407.1 GL888030 EGI69135.1 GBBI01003660 JAC15052.1 KQ434778 KZC04352.1 GDHC01001916 JAQ16713.1 GL435311 EFN73469.1 GGFK01010301 MBW43622.1 GECL01001062 JAP05062.1 ADTU01008925 KK107453 QOIP01000004 EZA50582.1 RLU23592.1 ACPB03026116 ACPB03026117 CH480820 EDW54109.1 KQ414572 KOC71313.1 GDIP01147644 LRGB01003123 JAJ75758.1 KZS04356.1 ADTU01008920 KQ435850 KOX70913.1 GDIP01031548 JAM72167.1 KQ978068 KYM97377.1 KZ288192 PBC34292.1 GL446459 EFN87770.1
KPJ15335.1 AGBW02007808 OWR54526.1 GAIX01012039 JAA80521.1 KZ150076 PZC73951.1 AJVK01001043 AAAB01008933 EAL40355.3 APCN01005565 JXUM01151776 KQ571296 KXJ68209.1 CH933807 EDW12999.1 ADMH02001479 ETN62390.1 CM000361 CM002910 EDX03580.1 KMY87828.1 GFDL01014674 JAV20371.1 GFDL01014671 JAV20374.1 CH963857 EDW76635.1 AXCN02001048 OUUW01000006 SPP82198.1 ATLV01023914 KE525347 KFB49900.1 CM000157 EDW87502.1 CH954177 EDV57587.1 AJ277122 AE014134 AY051884 AY060810 CH902667 EDV30613.1 CH379061 EAL32995.1 CH916368 EDW03844.1 CP012523 ALC38575.1 CH940649 EDW65085.1 UFQT01000279 SSX22705.1 JRES01000507 KNC30525.1 CVRI01000070 CRL07226.1 GECU01019521 JAS88185.1 GBXI01004002 JAD10290.1 GGFK01009544 MBW42865.1 GGFK01009485 MBW42806.1 GGFK01009545 MBW42866.1 GDHF01015186 JAI37128.1 GECZ01029008 JAS40761.1 GAKP01005928 JAC53024.1 CH477781 EAT36273.1 CH478692 EAT32652.1 CCAG010017571 GDHC01001319 JAQ17310.1 GBHO01025551 JAG18053.1 DS231930 EDS27256.1 GBRD01010415 JAG55409.1 NEVH01020337 PNF21657.1 KK852631 KDR19846.1 JXJN01018248 JXJN01023093 GAMC01010272 GAMC01010269 GAMC01010266 JAB96286.1 GECU01028321 JAS79385.1 GL767586 EFZ13407.1 GL888030 EGI69135.1 GBBI01003660 JAC15052.1 KQ434778 KZC04352.1 GDHC01001916 JAQ16713.1 GL435311 EFN73469.1 GGFK01010301 MBW43622.1 GECL01001062 JAP05062.1 ADTU01008925 KK107453 QOIP01000004 EZA50582.1 RLU23592.1 ACPB03026116 ACPB03026117 CH480820 EDW54109.1 KQ414572 KOC71313.1 GDIP01147644 LRGB01003123 JAJ75758.1 KZS04356.1 ADTU01008920 KQ435850 KOX70913.1 GDIP01031548 JAM72167.1 KQ978068 KYM97377.1 KZ288192 PBC34292.1 GL446459 EFN87770.1
Proteomes
UP000005204
UP000053240
UP000007151
UP000092462
UP000007062
UP000075840
+ More
UP000069940 UP000249989 UP000009192 UP000075903 UP000000673 UP000075900 UP000076408 UP000000304 UP000007798 UP000075882 UP000075886 UP000268350 UP000030765 UP000002282 UP000095300 UP000008711 UP000000803 UP000075880 UP000007801 UP000001819 UP000001070 UP000076407 UP000092553 UP000008792 UP000069272 UP000192221 UP000075902 UP000075881 UP000095301 UP000075885 UP000037069 UP000092445 UP000075920 UP000091820 UP000183832 UP000075901 UP000092443 UP000008820 UP000092444 UP000075884 UP000002320 UP000235965 UP000027135 UP000092460 UP000078200 UP000007755 UP000076502 UP000000311 UP000005205 UP000053097 UP000279307 UP000015103 UP000001292 UP000053825 UP000005203 UP000076858 UP000053105 UP000078542 UP000242457 UP000008237 UP000192223
UP000069940 UP000249989 UP000009192 UP000075903 UP000000673 UP000075900 UP000076408 UP000000304 UP000007798 UP000075882 UP000075886 UP000268350 UP000030765 UP000002282 UP000095300 UP000008711 UP000000803 UP000075880 UP000007801 UP000001819 UP000001070 UP000076407 UP000092553 UP000008792 UP000069272 UP000192221 UP000075902 UP000075881 UP000095301 UP000075885 UP000037069 UP000092445 UP000075920 UP000091820 UP000183832 UP000075901 UP000092443 UP000008820 UP000092444 UP000075884 UP000002320 UP000235965 UP000027135 UP000092460 UP000078200 UP000007755 UP000076502 UP000000311 UP000005205 UP000053097 UP000279307 UP000015103 UP000001292 UP000053825 UP000005203 UP000076858 UP000053105 UP000078542 UP000242457 UP000008237 UP000192223
Pfam
Interpro
SUPFAM
SSF63712
SSF63712
Gene 3D
ProteinModelPortal
H9JRI5
A0A0N0PDC5
A0A212FLB5
S4NP13
A0A2W1BI46
A0A1B0DJS2
+ More
Q5TSF5 A0A1Y9GLD9 A0A182G8A8 B4KEU9 A0A182VGJ1 W5JHK8 A0A182S510 A0A182YA38 B4Q941 A0A1Q3EYJ2 A0A1Q3EYJ3 B4MWZ6 A0A182LQ33 A0A182QUU8 A0A3B0KF10 A0A084WI56 B4NXE5 A0A1I8PBS7 B3NAR4 Q9VQM4 A0A182ITF6 B3N203 Q29L30 B4JAN6 A0A182X1U4 A0A0M4EP57 B4LU32 A0A182FV04 A0A1W4V7I2 A0A182TNC9 A0A182JRK8 A0A1I8N345 A0A182P8G8 A0A336LXP3 A0A0L0CE48 A0A1B0A4S1 A0A182VTC3 A0A1A9W6E8 A0A1J1J5J5 A0A1B6IMP2 A0A0A1XHB1 A0A2M4APZ2 A0A2M4APT8 A0A2M4APZ9 A0A182SYC7 A0A0K8VE29 A0A1B6ES60 A0A1A9YKG6 A0A034WEA8 Q16PH5 Q16EJ4 A0A1B0FC16 A0A146MFD4 A0A0A9XGY5 A0A182MYE1 B0WGW2 A0A0K8SQK4 A0A2J7PZ96 A0A1I8N346 A0A067R8H0 A0A1B0BPX4 A0A1A9USD2 W8BSM7 A0A1B6HXL7 E9J172 F4W9G0 A0A023F0N4 A0A154NXS3 A0A146MAD7 E1ZZ54 A0A2M4ASB2 A0A0V0GAW7 A0A1Y9GKN2 A0A158N984 A0A026W3D2 T1H9D3 B4I2W4 A0A1S4HEZ1 A0A0L7RKH2 A0A088AUJ5 A0A0P5FIR5 A0A158N982 A0A0M8ZWA3 A0A0P6ACI7 A0A195C978 A0A2A3ESN6 E2B964 A0A1W4XJL3
Q5TSF5 A0A1Y9GLD9 A0A182G8A8 B4KEU9 A0A182VGJ1 W5JHK8 A0A182S510 A0A182YA38 B4Q941 A0A1Q3EYJ2 A0A1Q3EYJ3 B4MWZ6 A0A182LQ33 A0A182QUU8 A0A3B0KF10 A0A084WI56 B4NXE5 A0A1I8PBS7 B3NAR4 Q9VQM4 A0A182ITF6 B3N203 Q29L30 B4JAN6 A0A182X1U4 A0A0M4EP57 B4LU32 A0A182FV04 A0A1W4V7I2 A0A182TNC9 A0A182JRK8 A0A1I8N345 A0A182P8G8 A0A336LXP3 A0A0L0CE48 A0A1B0A4S1 A0A182VTC3 A0A1A9W6E8 A0A1J1J5J5 A0A1B6IMP2 A0A0A1XHB1 A0A2M4APZ2 A0A2M4APT8 A0A2M4APZ9 A0A182SYC7 A0A0K8VE29 A0A1B6ES60 A0A1A9YKG6 A0A034WEA8 Q16PH5 Q16EJ4 A0A1B0FC16 A0A146MFD4 A0A0A9XGY5 A0A182MYE1 B0WGW2 A0A0K8SQK4 A0A2J7PZ96 A0A1I8N346 A0A067R8H0 A0A1B0BPX4 A0A1A9USD2 W8BSM7 A0A1B6HXL7 E9J172 F4W9G0 A0A023F0N4 A0A154NXS3 A0A146MAD7 E1ZZ54 A0A2M4ASB2 A0A0V0GAW7 A0A1Y9GKN2 A0A158N984 A0A026W3D2 T1H9D3 B4I2W4 A0A1S4HEZ1 A0A0L7RKH2 A0A088AUJ5 A0A0P5FIR5 A0A158N982 A0A0M8ZWA3 A0A0P6ACI7 A0A195C978 A0A2A3ESN6 E2B964 A0A1W4XJL3
PDB
5NWA
E-value=3.62306e-74,
Score=710
Ontologies
GO
PANTHER
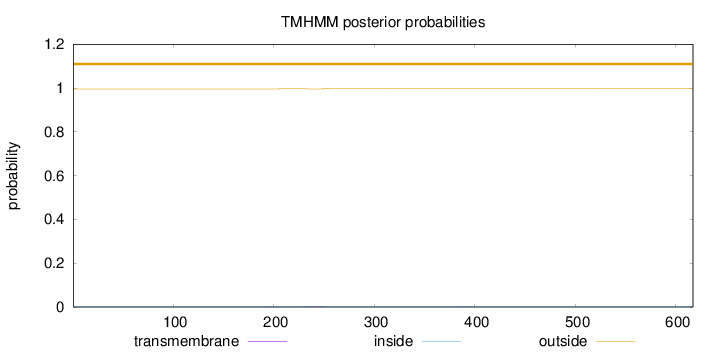
Topology
Subcellular location
Mitochondrion
Nucleus
Cytoplasm
Nucleus
Cytoplasm
Length:
617
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0610900000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00387
outside
1 - 617
Population Genetic Test Statistics
Pi
203.074771
Theta
180.04293
Tajima's D
0.733226
CLR
0.174142
CSRT
0.588220588970552
Interpretation
Uncertain
