Gene
KWMTBOMO14822
Pre Gene Modal
BGIBMGA013783
Annotation
AF461149_1_transposase_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.979
Sequence
CDS
ATGGAGTGGACGCTTAAAGAAGATCGTGTTGCAGTTATTGCGTTGCATCGTTGCGGTTACGCGCCAATTCAAATTTTTAACATACTGAAAAATTTGAATATAGCCAAAAGATTCGTTTATCGTACCATCAAACGATACAATGAAGACTCTAGTGTAGATGAGAGGTCAAGAAGTGGTCGCCCTCGGTCTGTTAGGACTCCAGAAGTGATAAAAGCTGTGAAGGCGCGAATTCAAAGAAATCCCAAACAAGAGAGCTACAACAAACAAAATGATAAGGTGTACGCACACAGTAGTGAAGAAGCGAGCAACCGTATTCCGCGTGTCCAACGAGGTCATTTTCCATCCTCGCTCATGGTATGGTTGGGAGTTTCTTATTGGGGCTTAACAGAGGTACATTTTTGTGAGAAAGGTGTAAAAACGAATGCAGTTGTGTATCAAAATACAGTCCTGACGAACCTTGTGGAACCTGTTTCTCATACCATGTTCAATAACAGGCACTGGGTATTCCAACAAGATTCGGCGCCAGCTCATAGAGCGAAGAGCACACAAGACTGGCTGGCGGCGCGTGAAATCGACTTCATCCGGCACGAAGACTGGCCATCCTCCAGTCCAGATTTGAATCCGTTAGATTACAAGATATGGCAACACTTGGAGGAAAAGGCGTGCTCAAAGCCTCATCCCAATTTGGAGTCACTCAAGACATCCTTGATTAAGGCAGCCGCCGATATTGACACGGACCTCGTTCGTGCTGCGATAGACGACTGGCCGCGCAGATTGAAGGCCTGTATTCAAAATCACGGAGGTCATTTTGAATAA
Protein
MEWTLKEDRVAVIALHRCGYAPIQIFNILKNLNIAKRFVYRTIKRYNEDSSVDERSRSGRPRSVRTPEVIKAVKARIQRNPKQESYNKQNDKVYAHSSEEASNRIPRVQRGHFPSSLMVWLGVSYWGLTEVHFCEKGVKTNAVVYQNTVLTNLVEPVSHTMFNNRHWVFQQDSAPAHRAKSTQDWLAAREIDFIRHEDWPSSSPDLNPLDYKIWQHLEEKACSKPHPNLESLKTSLIKAAADIDTDLVRAAIDDWPRRLKACIQNHGGHFE
Summary
Similarity
Belongs to the TGF-beta family.
Uniprot
EMBL
Proteomes
Interpro
Gene 3D
ProteinModelPortal
PDB
5CR4
E-value=7.99548e-10,
Score=151
Ontologies
GO
PANTHER
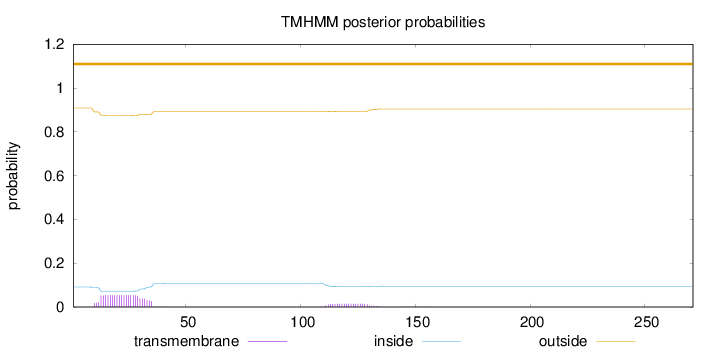
Topology
Subcellular location
Secreted
Length:
271
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.45266
Exp number, first 60 AAs:
1.16687
Total prob of N-in:
0.09124
outside
1 - 271
Population Genetic Test Statistics
Pi
36.580811
Theta
29.984722
Tajima's D
0.18324
CLR
0.109763
CSRT
0.427078646067697
Interpretation
Uncertain
