Pre Gene Modal
BGIBMGA012142
Annotation
PREDICTED:_mannosyl-oligosaccharide_glucosidase_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.025 Mitochondrial Reliability : 1.656
Sequence
CDS
ATGGTCAGACACAGAAAAGGCGGTAAAGAGCCGCATAAAAGTTCTAATAATGAAACCAGTAGTAGTTCTGGTTCCAGTGATACTTCGGTAGAAAAAAAACCATTTAAAATACTGTCTCTATGGAAAACTTTGGTCGGTTTTATATGTTTAGTAGTGGCCATATACGGCGGTGTGGTGGGTTACTTAGAGACGAGAGTAAACACGCCGTTTGACGACGTTAAGGTTGTACAAAAATCTGGTCTTGAGGTGCCTGATCGATATTGGGGCACATACAGACCTGGTGTATACTTAGGTATGAAAAGTCGGGAGCCATGGTCCCCAGTTTTTGGTCTAATGTGGTATGAGCTTGCTGCTGCTGCTCAAAAAGGAATCAGACACTGGTGCGATCAGGGTGACAACTTACCTACGTATGGCTGGCTCCGACATGATGGTGTGACTTTCGGAGAACAGCTGATATCGGACCCCCCTCATCAGATTGTTACCAGTTTCGTAAAGACACCGGGAGGGGCTCACGGAGGTCACTGGACTGCCAGGATCAATGTTACTGCTACGGCAAGATCACCTGGGCCCTTCGCGCTGGTATGGTACGGCGCTTTAGATGAATCCCTTGGACCTGCCGCCCCACATTCGAGGCTGTGGATTGAAGGAGGGGACATACTCGGCCACACATCGAATTTAAGGAATTTTCGCCTTAATCTAGTACCGCATAAAGGTAAAATTCTTCACAAGTCATTTTCGGAGGCGCACGCCGAAGGCCTCCATTTGCTCAAGGAGAAATTCTATGGCTTATTGAGGGGAGACAAGCATTCTAAAATCGGTAACATAGCCGTTTTGTCCGAAGACGACGACGTCATTGTCAAGAATAAGGACGCAAATTTCGTCCTAATACAATTGATAGTTGAAGCGCCGTTCGTTCTAGACGTAGTTTATACCACAGAGGAGCTACCGACTCCTCCACTTAAAGGAGACGAGTTCACCAAGAGTCTAGAAGAGAAGAAGAAGTCCTTCGACAATGAGTTCGAGAAGAAGTTTAAATTGCAAGAAAAGGGGTATCGAAAAGAAGATGTTGACGTGGCGAAAGCGGCGCTCTCAAATATGCTCGGCGGGATCGGGTACTTCTATGGCGCCGGCCGAGTTCAGTCCCAGTACACGAAGGAGTCGGTGCCGTACTGGCGGGCGCCCTTGTACACGGCCGTTCCGAGCAGATCATTCTTCCCTCGAGGATTCCTCTGGGACGAGGGCTTCCACGGACTTCTGATCGGAAGGTGGTCTCCGGATATTGAGCTCGATATAGCCGCTCATTGGCTGGACCTCATCAACGTGGAAGGCTGGATAGCGAGGGAACAGATTTTAGGCGTCGAAGCCTTAGCGAGAGTTCCCAAGGAGTTCGTAGTGCAGAACAACGCGGCGGCAAATCCTCCTATACTGTTAATAACACTGGCAAGGCTGGTGCAGCTGCGCCCTGAGCTGTTCAAGCCCCAGGGTCGGCTTAGGGGCACGCTCGACCGGATGTTCAGTAGACTGCAGGCCTGGTACCAGTGGTTCCAGACGACCCAAAAGGGAGAGGAGGCCACGTCGTACCGCTGGAGGGGTCGGGAGGACGACGGCCTCCAGCTCAACCCCAAAACCTTAACGTCCGGCCTCGACGATTATCCGAGGGCCTCGCATCCGTCAGACATCGAGCGCCATGTGGATCTTCGGTGTTGGATGTACGCGGCCGCCGACGCAATGGTCACAATAGCGGACGTCCTACAGCGTGACGTCAGCAAGTTCGAAACGACAAGGGATCAGTTGGGCGACGAAGATCTGCTCAACGAACTCCACTGGTCGACTCGCTCGCAAACCTACGCTGACTACGGTCTCCACACGGACGGAGTGCGCCTCGTCAGACAGCAGTCCAAGAAGCCGAACGAGAACGGTAAAATGGTCCGAGTCGTGACCACCGCGCCGCAACCGATGCTCGTCACGTCCGCTTTCGGATACATATCCTTGTTCCCGATGCTGCTGAAGGTCCTTAAATCTGACAGCGAGAAACTGGAGAGAATACTGGATTCTCTCGATAAGCCCGACTTATTGTGGTCTCCGTACGGACTTAGATCGCTGTCAAAATCATCGCCATTATACATGAAGAGGAACACGGAACACGATCCGCCATATTGGCGCGGGCAAATATGGATGAACATTAATTATTTAGCGCTTTCGAGCTTGAAGTACTACGCGACCGTGCCCGGCCCGTACAAGGCGAGAGCGGGACAGCTGCACGATCGTCTGAGAGAGAACGTCGTCCGCAACATCATATCCGAGTACAGGAGGACGGGCTACCTCTGGGAGCAGTACTCTGGCGAAGATGGCAAGGGCTCTGGATGTAGACCGTTCAGCGGCTGGACTGGACTCTTGGTTCTCATCATGGCTGAACAGTATTAG
Protein
MVRHRKGGKEPHKSSNNETSSSSGSSDTSVEKKPFKILSLWKTLVGFICLVVAIYGGVVGYLETRVNTPFDDVKVVQKSGLEVPDRYWGTYRPGVYLGMKSREPWSPVFGLMWYELAAAAQKGIRHWCDQGDNLPTYGWLRHDGVTFGEQLISDPPHQIVTSFVKTPGGAHGGHWTARINVTATARSPGPFALVWYGALDESLGPAAPHSRLWIEGGDILGHTSNLRNFRLNLVPHKGKILHKSFSEAHAEGLHLLKEKFYGLLRGDKHSKIGNIAVLSEDDDVIVKNKDANFVLIQLIVEAPFVLDVVYTTEELPTPPLKGDEFTKSLEEKKKSFDNEFEKKFKLQEKGYRKEDVDVAKAALSNMLGGIGYFYGAGRVQSQYTKESVPYWRAPLYTAVPSRSFFPRGFLWDEGFHGLLIGRWSPDIELDIAAHWLDLINVEGWIAREQILGVEALARVPKEFVVQNNAAANPPILLITLARLVQLRPELFKPQGRLRGTLDRMFSRLQAWYQWFQTTQKGEEATSYRWRGREDDGLQLNPKTLTSGLDDYPRASHPSDIERHVDLRCWMYAAADAMVTIADVLQRDVSKFETTRDQLGDEDLLNELHWSTRSQTYADYGLHTDGVRLVRQQSKKPNENGKMVRVVTTAPQPMLVTSAFGYISLFPMLLKVLKSDSEKLERILDSLDKPDLLWSPYGLRSLSKSSPLYMKRNTEHDPPYWRGQIWMNINYLALSSLKYYATVPGPYKARAGQLHDRLRENVVRNIISEYRRTGYLWEQYSGEDGKGSGCRPFSGWTGLLVLIMAEQY
Summary
Uniprot
A0A2A4K7J3
A0A212F8S0
A0A3S2NID8
A0A2W1BJZ2
A0A194Q146
A0A1W4WJ19
+ More
A0A1W4W7F2 A0A1L8E1B7 A0A1L8E1C8 A0A0A1X2Q2 A0A1B0DQ79 A0A034W9H9 N6U0V5 A0A2P8XLS8 A0A0L0C2N1 A0A1I8NIB1 A0A3B0KRF5 A0A139WB11 T1PNQ3 B5DP06 B4HA11 A0A1I8PCG0 T1PL70 A0A1Y1KVM4 B4R2T0 B4PYC7 A0A0K8VSF7 W8AWU4 A0A2J7RH57 A0A0K8TMY1 A9UNC0 Q95R68 Q9VZ04 A0A1W4UNQ4 B3NV78 A0A0M5JAC2 A0A067R1F9 B3N097 A0A1J1IJM4 A0A1B0FQ35 B4M1C2 A0A1A9UT33 T1HWP8 A0A1A9ZF48 A0A1A9WAU0 A0A1A9XLW7 B4L3C3 A0A2P2I7M2 B4JNW2 B4NDC9 A0A224XJP7 A0A069DWI0 E2A6V1 A0A154P222 A0A023EYB7 A0A0L7RJJ7 K7J9E2 A0A158NKH2 A0A195F663 A0A1B6DLQ2 A0A3Q0JAS1 A0A195BV50 E0VYK6 A0A0C9R244 F4X1L8 E2C3D1 A0A151WTE3 A0A2A3EDP0 A0A0N0BF94 A0A088ADG3 A0A310STE4 A0A1B6GGS0 A0A2R7VNR6 B4IE08 A0A195CYT6 A0A195EJ74 A0A026WVH2 A0A3L8DQH4 A0A3R7Q9X2 A0A162D7A7 A0A0P6FY28 E9G6K4 U5ET53 A0A0N8CSY3 A0A0P5NMH9 A0A0P6AA82 A0A0P5ZMN1 A0A0P5Q893 A0A0P5PLS4 A0A0P5RIP2 A0A293L679 A0A226DJA8 A0A2G8L5C7 T1ISH0 A0A1Z5L5Y4 A0A2R5LFR3 K1PZ89 C3ZIN6 A0A1S3HWL5
A0A1W4W7F2 A0A1L8E1B7 A0A1L8E1C8 A0A0A1X2Q2 A0A1B0DQ79 A0A034W9H9 N6U0V5 A0A2P8XLS8 A0A0L0C2N1 A0A1I8NIB1 A0A3B0KRF5 A0A139WB11 T1PNQ3 B5DP06 B4HA11 A0A1I8PCG0 T1PL70 A0A1Y1KVM4 B4R2T0 B4PYC7 A0A0K8VSF7 W8AWU4 A0A2J7RH57 A0A0K8TMY1 A9UNC0 Q95R68 Q9VZ04 A0A1W4UNQ4 B3NV78 A0A0M5JAC2 A0A067R1F9 B3N097 A0A1J1IJM4 A0A1B0FQ35 B4M1C2 A0A1A9UT33 T1HWP8 A0A1A9ZF48 A0A1A9WAU0 A0A1A9XLW7 B4L3C3 A0A2P2I7M2 B4JNW2 B4NDC9 A0A224XJP7 A0A069DWI0 E2A6V1 A0A154P222 A0A023EYB7 A0A0L7RJJ7 K7J9E2 A0A158NKH2 A0A195F663 A0A1B6DLQ2 A0A3Q0JAS1 A0A195BV50 E0VYK6 A0A0C9R244 F4X1L8 E2C3D1 A0A151WTE3 A0A2A3EDP0 A0A0N0BF94 A0A088ADG3 A0A310STE4 A0A1B6GGS0 A0A2R7VNR6 B4IE08 A0A195CYT6 A0A195EJ74 A0A026WVH2 A0A3L8DQH4 A0A3R7Q9X2 A0A162D7A7 A0A0P6FY28 E9G6K4 U5ET53 A0A0N8CSY3 A0A0P5NMH9 A0A0P6AA82 A0A0P5ZMN1 A0A0P5Q893 A0A0P5PLS4 A0A0P5RIP2 A0A293L679 A0A226DJA8 A0A2G8L5C7 T1ISH0 A0A1Z5L5Y4 A0A2R5LFR3 K1PZ89 C3ZIN6 A0A1S3HWL5
Pubmed
22118469
28756777
26354079
25830018
25348373
23537049
+ More
29403074 26108605 25315136 18362917 19820115 15632085 17994087 28004739 17550304 24495485 26369729 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24845553 26334808 20798317 25474469 20075255 21347285 20566863 21719571 24508170 30249741 21292972 29023486 28528879 22992520 18563158
29403074 26108605 25315136 18362917 19820115 15632085 17994087 28004739 17550304 24495485 26369729 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24845553 26334808 20798317 25474469 20075255 21347285 20566863 21719571 24508170 30249741 21292972 29023486 28528879 22992520 18563158
EMBL
NWSH01000049
PCG80207.1
AGBW02009680
OWR50146.1
RSAL01000084
RVE48369.1
+ More
KZ150076 PZC73964.1 KQ459580 KPI99276.1 GFDF01001597 JAV12487.1 GFDF01001595 JAV12489.1 GBXI01009262 JAD05030.1 AJVK01018925 AJVK01018926 GAKP01008504 JAC50448.1 APGK01053965 KB741239 KB632275 ENN72177.1 ERL91145.1 PYGN01001763 PSN32948.1 JRES01000984 KNC26516.1 OUUW01000011 SPP86498.1 KQ971379 KYB25099.1 KA649720 AFP64349.1 CH379066 EDY73258.1 CH479235 EDW36679.1 KA649471 KA649472 AFP64100.1 GEZM01075181 JAV64220.1 CM000366 EDX17629.1 CM000162 EDX01989.2 GDHF01010493 JAI41821.1 GAMC01015983 JAB90572.1 NEVH01003746 PNF40173.1 GDAI01001881 JAI15722.1 BT031284 ABY20525.1 AY061591 AAL29139.1 AE014298 AAF48028.4 AFH07335.1 CH954180 EDV46066.1 CP012528 ALC48411.1 KK853097 KDR11423.1 CH902640 EDV38301.1 CVRI01000054 CRL00392.1 CCAG010006700 CH940651 EDW65476.1 ACPB03026615 CH933810 EDW07051.1 IACF01004429 LAB70023.1 CH916371 EDV92405.1 CH964239 EDW82838.1 GFTR01008073 JAW08353.1 GBGD01000501 JAC88388.1 GL437203 EFN70817.1 KQ434803 KZC05985.1 GBBI01004392 JAC14320.1 KQ414581 KOC71009.1 ADTU01002066 ADTU01002067 KQ981805 KYN35554.1 GEDC01010685 JAS26613.1 KQ976403 KYM92130.1 DS235845 EEB18462.1 GBYB01006907 GBYB01006908 GBYB01010634 JAG76674.1 JAG76675.1 JAG80401.1 GL888531 EGI59676.1 GL452320 EFN77469.1 KQ982757 KYQ51106.1 KZ288271 PBC29828.1 KQ435812 KOX72748.1 KQ760549 OAD59879.1 GECZ01008130 JAS61639.1 KK854008 PTY09183.1 CH480830 EDW45816.1 KQ977110 KYN05737.1 KQ978801 KYN28303.1 KK107102 EZA59646.1 QOIP01000005 RLU22685.1 QCYY01002168 ROT72368.1 LRGB01002384 KZS07904.1 GDIQ01041305 JAN53432.1 GL732533 EFX84976.1 GANO01002082 JAB57789.1 GDIP01101983 JAM01732.1 GDIQ01140262 JAL11464.1 GDIP01033089 JAM70626.1 GDIP01042680 JAM61035.1 GDIQ01117824 JAL33902.1 GDIQ01147820 JAL03906.1 GDIQ01119413 JAL32313.1 GFWV01002767 MAA27497.1 LNIX01000017 OXA45622.1 MRZV01000214 PIK55466.1 JH431432 GFJQ02004290 JAW02680.1 GGLE01004183 MBY08309.1 JH816673 EKC26958.1 GG666628 EEN47571.1
KZ150076 PZC73964.1 KQ459580 KPI99276.1 GFDF01001597 JAV12487.1 GFDF01001595 JAV12489.1 GBXI01009262 JAD05030.1 AJVK01018925 AJVK01018926 GAKP01008504 JAC50448.1 APGK01053965 KB741239 KB632275 ENN72177.1 ERL91145.1 PYGN01001763 PSN32948.1 JRES01000984 KNC26516.1 OUUW01000011 SPP86498.1 KQ971379 KYB25099.1 KA649720 AFP64349.1 CH379066 EDY73258.1 CH479235 EDW36679.1 KA649471 KA649472 AFP64100.1 GEZM01075181 JAV64220.1 CM000366 EDX17629.1 CM000162 EDX01989.2 GDHF01010493 JAI41821.1 GAMC01015983 JAB90572.1 NEVH01003746 PNF40173.1 GDAI01001881 JAI15722.1 BT031284 ABY20525.1 AY061591 AAL29139.1 AE014298 AAF48028.4 AFH07335.1 CH954180 EDV46066.1 CP012528 ALC48411.1 KK853097 KDR11423.1 CH902640 EDV38301.1 CVRI01000054 CRL00392.1 CCAG010006700 CH940651 EDW65476.1 ACPB03026615 CH933810 EDW07051.1 IACF01004429 LAB70023.1 CH916371 EDV92405.1 CH964239 EDW82838.1 GFTR01008073 JAW08353.1 GBGD01000501 JAC88388.1 GL437203 EFN70817.1 KQ434803 KZC05985.1 GBBI01004392 JAC14320.1 KQ414581 KOC71009.1 ADTU01002066 ADTU01002067 KQ981805 KYN35554.1 GEDC01010685 JAS26613.1 KQ976403 KYM92130.1 DS235845 EEB18462.1 GBYB01006907 GBYB01006908 GBYB01010634 JAG76674.1 JAG76675.1 JAG80401.1 GL888531 EGI59676.1 GL452320 EFN77469.1 KQ982757 KYQ51106.1 KZ288271 PBC29828.1 KQ435812 KOX72748.1 KQ760549 OAD59879.1 GECZ01008130 JAS61639.1 KK854008 PTY09183.1 CH480830 EDW45816.1 KQ977110 KYN05737.1 KQ978801 KYN28303.1 KK107102 EZA59646.1 QOIP01000005 RLU22685.1 QCYY01002168 ROT72368.1 LRGB01002384 KZS07904.1 GDIQ01041305 JAN53432.1 GL732533 EFX84976.1 GANO01002082 JAB57789.1 GDIP01101983 JAM01732.1 GDIQ01140262 JAL11464.1 GDIP01033089 JAM70626.1 GDIP01042680 JAM61035.1 GDIQ01117824 JAL33902.1 GDIQ01147820 JAL03906.1 GDIQ01119413 JAL32313.1 GFWV01002767 MAA27497.1 LNIX01000017 OXA45622.1 MRZV01000214 PIK55466.1 JH431432 GFJQ02004290 JAW02680.1 GGLE01004183 MBY08309.1 JH816673 EKC26958.1 GG666628 EEN47571.1
Proteomes
UP000218220
UP000007151
UP000283053
UP000053268
UP000192223
UP000092462
+ More
UP000019118 UP000030742 UP000245037 UP000037069 UP000095301 UP000268350 UP000007266 UP000001819 UP000008744 UP000095300 UP000000304 UP000002282 UP000235965 UP000000803 UP000192221 UP000008711 UP000092553 UP000027135 UP000007801 UP000183832 UP000092444 UP000008792 UP000078200 UP000015103 UP000092445 UP000091820 UP000092443 UP000009192 UP000001070 UP000007798 UP000000311 UP000076502 UP000053825 UP000002358 UP000005205 UP000078541 UP000079169 UP000078540 UP000009046 UP000007755 UP000008237 UP000075809 UP000242457 UP000053105 UP000005203 UP000001292 UP000078542 UP000078492 UP000053097 UP000279307 UP000283509 UP000076858 UP000000305 UP000198287 UP000230750 UP000005408 UP000001554 UP000085678
UP000019118 UP000030742 UP000245037 UP000037069 UP000095301 UP000268350 UP000007266 UP000001819 UP000008744 UP000095300 UP000000304 UP000002282 UP000235965 UP000000803 UP000192221 UP000008711 UP000092553 UP000027135 UP000007801 UP000183832 UP000092444 UP000008792 UP000078200 UP000015103 UP000092445 UP000091820 UP000092443 UP000009192 UP000001070 UP000007798 UP000000311 UP000076502 UP000053825 UP000002358 UP000005205 UP000078541 UP000079169 UP000078540 UP000009046 UP000007755 UP000008237 UP000075809 UP000242457 UP000053105 UP000005203 UP000001292 UP000078542 UP000078492 UP000053097 UP000279307 UP000283509 UP000076858 UP000000305 UP000198287 UP000230750 UP000005408 UP000001554 UP000085678
Interpro
Gene 3D
ProteinModelPortal
A0A2A4K7J3
A0A212F8S0
A0A3S2NID8
A0A2W1BJZ2
A0A194Q146
A0A1W4WJ19
+ More
A0A1W4W7F2 A0A1L8E1B7 A0A1L8E1C8 A0A0A1X2Q2 A0A1B0DQ79 A0A034W9H9 N6U0V5 A0A2P8XLS8 A0A0L0C2N1 A0A1I8NIB1 A0A3B0KRF5 A0A139WB11 T1PNQ3 B5DP06 B4HA11 A0A1I8PCG0 T1PL70 A0A1Y1KVM4 B4R2T0 B4PYC7 A0A0K8VSF7 W8AWU4 A0A2J7RH57 A0A0K8TMY1 A9UNC0 Q95R68 Q9VZ04 A0A1W4UNQ4 B3NV78 A0A0M5JAC2 A0A067R1F9 B3N097 A0A1J1IJM4 A0A1B0FQ35 B4M1C2 A0A1A9UT33 T1HWP8 A0A1A9ZF48 A0A1A9WAU0 A0A1A9XLW7 B4L3C3 A0A2P2I7M2 B4JNW2 B4NDC9 A0A224XJP7 A0A069DWI0 E2A6V1 A0A154P222 A0A023EYB7 A0A0L7RJJ7 K7J9E2 A0A158NKH2 A0A195F663 A0A1B6DLQ2 A0A3Q0JAS1 A0A195BV50 E0VYK6 A0A0C9R244 F4X1L8 E2C3D1 A0A151WTE3 A0A2A3EDP0 A0A0N0BF94 A0A088ADG3 A0A310STE4 A0A1B6GGS0 A0A2R7VNR6 B4IE08 A0A195CYT6 A0A195EJ74 A0A026WVH2 A0A3L8DQH4 A0A3R7Q9X2 A0A162D7A7 A0A0P6FY28 E9G6K4 U5ET53 A0A0N8CSY3 A0A0P5NMH9 A0A0P6AA82 A0A0P5ZMN1 A0A0P5Q893 A0A0P5PLS4 A0A0P5RIP2 A0A293L679 A0A226DJA8 A0A2G8L5C7 T1ISH0 A0A1Z5L5Y4 A0A2R5LFR3 K1PZ89 C3ZIN6 A0A1S3HWL5
A0A1W4W7F2 A0A1L8E1B7 A0A1L8E1C8 A0A0A1X2Q2 A0A1B0DQ79 A0A034W9H9 N6U0V5 A0A2P8XLS8 A0A0L0C2N1 A0A1I8NIB1 A0A3B0KRF5 A0A139WB11 T1PNQ3 B5DP06 B4HA11 A0A1I8PCG0 T1PL70 A0A1Y1KVM4 B4R2T0 B4PYC7 A0A0K8VSF7 W8AWU4 A0A2J7RH57 A0A0K8TMY1 A9UNC0 Q95R68 Q9VZ04 A0A1W4UNQ4 B3NV78 A0A0M5JAC2 A0A067R1F9 B3N097 A0A1J1IJM4 A0A1B0FQ35 B4M1C2 A0A1A9UT33 T1HWP8 A0A1A9ZF48 A0A1A9WAU0 A0A1A9XLW7 B4L3C3 A0A2P2I7M2 B4JNW2 B4NDC9 A0A224XJP7 A0A069DWI0 E2A6V1 A0A154P222 A0A023EYB7 A0A0L7RJJ7 K7J9E2 A0A158NKH2 A0A195F663 A0A1B6DLQ2 A0A3Q0JAS1 A0A195BV50 E0VYK6 A0A0C9R244 F4X1L8 E2C3D1 A0A151WTE3 A0A2A3EDP0 A0A0N0BF94 A0A088ADG3 A0A310STE4 A0A1B6GGS0 A0A2R7VNR6 B4IE08 A0A195CYT6 A0A195EJ74 A0A026WVH2 A0A3L8DQH4 A0A3R7Q9X2 A0A162D7A7 A0A0P6FY28 E9G6K4 U5ET53 A0A0N8CSY3 A0A0P5NMH9 A0A0P6AA82 A0A0P5ZMN1 A0A0P5Q893 A0A0P5PLS4 A0A0P5RIP2 A0A293L679 A0A226DJA8 A0A2G8L5C7 T1ISH0 A0A1Z5L5Y4 A0A2R5LFR3 K1PZ89 C3ZIN6 A0A1S3HWL5
PDB
5MHF
E-value=7.27518e-153,
Score=1389
Ontologies
PATHWAY
GO
PANTHER
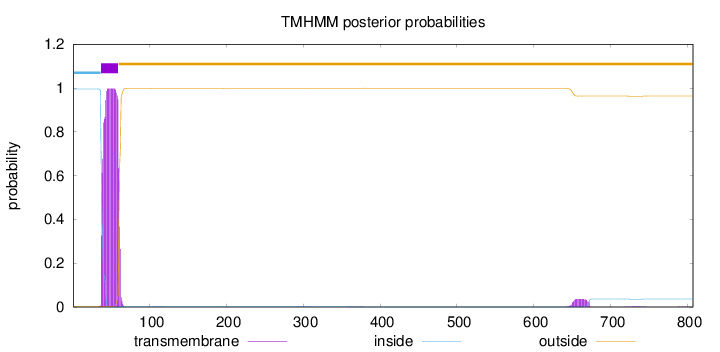
Topology
Length:
807
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.18217
Exp number, first 60 AAs:
21.61244
Total prob of N-in:
0.99614
POSSIBLE N-term signal
sequence
inside
1 - 36
TMhelix
37 - 59
outside
60 - 807
Population Genetic Test Statistics
Pi
262.445073
Theta
154.579213
Tajima's D
1.502709
CLR
0.89237
CSRT
0.786210689465527
Interpretation
Uncertain
