Gene
KWMTBOMO14816
Pre Gene Modal
BGIBMGA012202
Annotation
PREDICTED:_LIM_domain-binding_protein_2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.42
Sequence
CDS
ATGCCAGTGCTGCTCGAAGTCTGCGTACACCCCGAAAGCGCTAGATGGCGCGGTGACAAAGTGGCCTACATGCCGGTGGCGTTGGGCTCGCTCGGCGGCGAGGGCGAGTACCGCGACTACGGCCATCATCTATCGCCGCATCATACGCCGCATCCGCCGCACATACAGTACCATGAGTACGCGCCGCCGCCGCCGCCGCCGATCTACCACCCGATACCGGACCCCTACTTCAGGCGGCACACGCCATACTTCGGGCAACCAGATTACAGAGTATATGAACTCAATAAGAGGTTACAGCAGAGGACGGAGGACTCTGACAACCTCTGGTGGGACGCGTTCGCGACGGAATTCTTCGAGGATGACGCGACGCTAACACTAACATTCTGTTTAGAAGACGGACCCAAAAGATACACGATAGGGAGGACCCTGATACCTCGCTACTTCCGCAGTATATACGAAGGCGGCGTCTCAGAGTTGTACTACACGATGCGCCAGCCCAAAGAGTCCTTCCACAACACGAGCATCACGCTGGACTGCGATCATTGCACAATGGTCACGCACCACGGCAAGCCTATGTTCACGAAGGTGTGCACGGAAGGCAGACTGATCCTCGAGTTCACGTTCGACGACCTCATGCGGATCAAGTCGTGGCACATGGCGGTTCGCGCGCACCGCGAGCTGATCCCGCGGCAGGCGGTCCACCCGCCGGACCACGCCGCGCTCGACCAGCTCACGAAGAACATCACACGGCAGGGCATCACGAATTCAACGCTTAACTACTTAAGGTTATGCGTGATATTGGAGCCGATGCAAGAGCTCATGTCGCGGCACAAAGCGTACGCGCTGTCGCCGCGGGACTGTCTAAAGACAACGCTCTTCCAGAAATGGCAGCGGATGGTCGCACCTCCTGAGTCCCAGCGGCCGGCGAGTAAGCGACGTAAGCGCAAAGGCAGTGCGGGCGCTAATGCGGCGCCGCCCGCGCCCGCCAAGAAACGATCACCCGGACCTAACTTCAGCCTCGCCTCCCAGGACGTGATGGTAGTAGGCGAACCGTCGCTGATGGGCGGCGAGTTCGGCGACGAGGACGAGCGGCTGATCACGCGGCTGGAGAACACGCAGTACGAGGGCGAGGGCGTGGAGTGGAGCGCCCCGCCCCCCGCGTCCCCCGCCAAGACCCCCCCCGGCACCCACTGA
Protein
MPVLLEVCVHPESARWRGDKVAYMPVALGSLGGEGEYRDYGHHLSPHHTPHPPHIQYHEYAPPPPPPIYHPIPDPYFRRHTPYFGQPDYRVYELNKRLQQRTEDSDNLWWDAFATEFFEDDATLTLTFCLEDGPKRYTIGRTLIPRYFRSIYEGGVSELYYTMRQPKESFHNTSITLDCDHCTMVTHHGKPMFTKVCTEGRLILEFTFDDLMRIKSWHMAVRAHRELIPRQAVHPPDHAALDQLTKNITRQGITNSTLNYLRLCVILEPMQELMSRHKAYALSPRDCLKTTLFQKWQRMVAPPESQRPASKRRKRKGSAGANAAPPAPAKKRSPGPNFSLASQDVMVVGEPSLMGGEFGDEDERLITRLENTQYEGEGVEWSAPPPASPAKTPPGTH
Summary
Uniprot
U6C886
A0A194Q2Q2
H9JRP2
A0A212EWG3
A0A2A4K8F4
U6C771
+ More
S4PMP0 A0A2A4K818 A0A0N1PHD8 A0A1B6DPS9 A0A3Q0IRJ0 A0A1B6E8X5 A0A0V0GCW0 A0A146LE46 V5G3G0 A0A1Y1LQU4 A0A1Y1LNI5 A0A1B6F6X4 A0A0A9WWL7 D6WBT5 A0A023F3G4 A0A1B6CIC4 A0A146L0J9 A0A1B6DFI6 A0A1W4XKX9 A0A1Y1LSE5 E0VPP1 T1IZ74 A0A224XQP1 A0A146LV70 A0A088ALZ2 A0A069DUT5 A0A0A9X6Z6 A0A154PK00 A0A2D3E2V8 A0A2R7WFT9 A0A310SES3 B7Q6T8 A0A2J7QQY3 A0A1J1IZU8 A0A0P4W827 K7JAU8 A0A2M4AAX5 A0A2M4CNX0 A0A182GN90 A0A2M4ABK6 A0A2M4AC57 A0A1B0D887 A0A1J1J010 A0A2M4AB23 A0A2M4BHK3 A0A182JSI6 A0A2M4BHP5 A0A182RJV7 A0A2M3Z261 A0A2M4CPR3 A0A1Y1LNJ8 A0A2M4CNH1 A0A2M4BH86 A0A1Q3FUE9 V5HAI1 Q16K77 Q16R65 A0A0C9Q9J5 A0A0P5G457 A0A067QIH5 A0A1L8DHC6 A0A084W172 A0A2M4AJM5 A0A182PMS5 A0A2M4BHJ7 A0A0C9RTY2 A0A1J1J2J0 A0A1D2MJD4 A0A0P4ZPX5 A0A0N8AIP9 A0A0C9R417 A0A2M4CNM3 A0A2M3ZGI4 Q7PMR9 A0A087U0U1 A0A2R5LK47 A0A293N4P5 A0A1Z5LGR6 A0A0P5BNK7 A0A0P5BIW9 E9FX36 A0A1B0CUW2 A0A0P5CII5 A0A087T8P6 U4UUQ9 A0A182WAX3 A0A182J471 A0A0P5DW26 V5IHZ1 A0A131Y3I2 A0A1E1XBB8 A0A0P6BQG8 A0A0P6HQS8
S4PMP0 A0A2A4K818 A0A0N1PHD8 A0A1B6DPS9 A0A3Q0IRJ0 A0A1B6E8X5 A0A0V0GCW0 A0A146LE46 V5G3G0 A0A1Y1LQU4 A0A1Y1LNI5 A0A1B6F6X4 A0A0A9WWL7 D6WBT5 A0A023F3G4 A0A1B6CIC4 A0A146L0J9 A0A1B6DFI6 A0A1W4XKX9 A0A1Y1LSE5 E0VPP1 T1IZ74 A0A224XQP1 A0A146LV70 A0A088ALZ2 A0A069DUT5 A0A0A9X6Z6 A0A154PK00 A0A2D3E2V8 A0A2R7WFT9 A0A310SES3 B7Q6T8 A0A2J7QQY3 A0A1J1IZU8 A0A0P4W827 K7JAU8 A0A2M4AAX5 A0A2M4CNX0 A0A182GN90 A0A2M4ABK6 A0A2M4AC57 A0A1B0D887 A0A1J1J010 A0A2M4AB23 A0A2M4BHK3 A0A182JSI6 A0A2M4BHP5 A0A182RJV7 A0A2M3Z261 A0A2M4CPR3 A0A1Y1LNJ8 A0A2M4CNH1 A0A2M4BH86 A0A1Q3FUE9 V5HAI1 Q16K77 Q16R65 A0A0C9Q9J5 A0A0P5G457 A0A067QIH5 A0A1L8DHC6 A0A084W172 A0A2M4AJM5 A0A182PMS5 A0A2M4BHJ7 A0A0C9RTY2 A0A1J1J2J0 A0A1D2MJD4 A0A0P4ZPX5 A0A0N8AIP9 A0A0C9R417 A0A2M4CNM3 A0A2M3ZGI4 Q7PMR9 A0A087U0U1 A0A2R5LK47 A0A293N4P5 A0A1Z5LGR6 A0A0P5BNK7 A0A0P5BIW9 E9FX36 A0A1B0CUW2 A0A0P5CII5 A0A087T8P6 U4UUQ9 A0A182WAX3 A0A182J471 A0A0P5DW26 V5IHZ1 A0A131Y3I2 A0A1E1XBB8 A0A0P6BQG8 A0A0P6HQS8
Pubmed
EMBL
AB687554
BAO05323.1
KQ459580
KPI99279.1
BABH01031875
BABH01031876
+ More
BABH01031877 AGBW02011995 OWR45823.1 NWSH01000049 PCG80208.1 AB687555 BAO05324.1 GAIX01003455 JAA89105.1 PCG80209.1 KQ460396 KPJ15348.1 GEDC01009612 JAS27686.1 GEDC01002917 JAS34381.1 GECL01000387 JAP05737.1 GDHC01012188 JAQ06441.1 GALX01003906 JAB64560.1 GEZM01051083 JAV75198.1 GEZM01051078 JAV75202.1 GECZ01023819 JAS45950.1 GBHO01030742 JAG12862.1 KQ971314 EEZ97869.2 GBBI01002896 JAC15816.1 GEDC01024099 JAS13199.1 GDHC01017130 JAQ01499.1 GEDC01012916 JAS24382.1 GEZM01051080 JAV75200.1 AAZO01004210 DS235366 EEB15347.1 AFFK01020413 GFTR01006137 JAW10289.1 GDHC01008369 JAQ10260.1 GBGD01001189 JAC87700.1 GBHO01030744 JAG12860.1 KQ434938 KZC12172.1 MF140471 ATU31736.1 KK854718 PTY18131.1 KQ760561 OAD59784.1 ABJB010509821 ABJB010794332 DS870229 EEC14560.1 NEVH01012082 PNF30995.1 CVRI01000065 CRL05663.1 GDRN01065843 JAI64633.1 AAZX01002733 AAZX01007004 AAZX01008610 GGFK01004598 MBW37919.1 GGFL01002703 MBW66881.1 JXUM01075938 KQ562914 KXJ74855.1 GGFK01004677 MBW37998.1 GGFK01005054 MBW38375.1 AJVK01012568 AJVK01012569 AJVK01012570 AJVK01012571 CRL05664.1 GGFK01004664 MBW37985.1 GGFJ01003394 MBW52535.1 GGFJ01003372 MBW52513.1 GGFM01001842 MBW22593.1 GGFL01002700 MBW66878.1 GEZM01051081 JAV75199.1 GGFL01002686 MBW66864.1 GGFJ01003279 MBW52420.1 GFDL01003963 JAV31082.1 GANP01004098 JAB80370.1 CH477974 EAT34689.1 CH477723 EAT36891.1 GBYB01011048 JAG80815.1 GDIQ01246721 JAK05004.1 KK853313 KDR08625.1 GFDF01008225 JAV05859.1 ATLV01019238 KE525266 KFB43966.1 GGFK01007626 MBW40947.1 GGFJ01003353 MBW52494.1 GBYB01011046 JAG80813.1 CRL05662.1 LJIJ01001104 ODM92975.1 GDIP01214029 JAJ09373.1 GDIP01143545 JAJ79857.1 GBYB01011044 JAG80811.1 GGFL01002681 MBW66859.1 GGFM01006850 MBW27601.1 AAAB01008968 EAA13295.5 KK117621 KFM70980.1 GGLE01005776 MBY09902.1 GFWV01022999 MAA47726.1 GFJQ02000470 JAW06500.1 GDIP01187030 JAJ36372.1 GDIP01189224 JAJ34178.1 GL732526 EFX88004.1 AJWK01029714 GDIP01170066 JAJ53336.1 KK113976 KFM61485.1 KB632375 ERL93890.1 AXCP01007737 GDIP01151823 JAJ71579.1 GANP01004097 JAB80371.1 GEFM01002764 JAP73032.1 GFAC01002641 JAT96547.1 GDIP01012925 JAM90790.1 GDIQ01015549 JAN79188.1
BABH01031877 AGBW02011995 OWR45823.1 NWSH01000049 PCG80208.1 AB687555 BAO05324.1 GAIX01003455 JAA89105.1 PCG80209.1 KQ460396 KPJ15348.1 GEDC01009612 JAS27686.1 GEDC01002917 JAS34381.1 GECL01000387 JAP05737.1 GDHC01012188 JAQ06441.1 GALX01003906 JAB64560.1 GEZM01051083 JAV75198.1 GEZM01051078 JAV75202.1 GECZ01023819 JAS45950.1 GBHO01030742 JAG12862.1 KQ971314 EEZ97869.2 GBBI01002896 JAC15816.1 GEDC01024099 JAS13199.1 GDHC01017130 JAQ01499.1 GEDC01012916 JAS24382.1 GEZM01051080 JAV75200.1 AAZO01004210 DS235366 EEB15347.1 AFFK01020413 GFTR01006137 JAW10289.1 GDHC01008369 JAQ10260.1 GBGD01001189 JAC87700.1 GBHO01030744 JAG12860.1 KQ434938 KZC12172.1 MF140471 ATU31736.1 KK854718 PTY18131.1 KQ760561 OAD59784.1 ABJB010509821 ABJB010794332 DS870229 EEC14560.1 NEVH01012082 PNF30995.1 CVRI01000065 CRL05663.1 GDRN01065843 JAI64633.1 AAZX01002733 AAZX01007004 AAZX01008610 GGFK01004598 MBW37919.1 GGFL01002703 MBW66881.1 JXUM01075938 KQ562914 KXJ74855.1 GGFK01004677 MBW37998.1 GGFK01005054 MBW38375.1 AJVK01012568 AJVK01012569 AJVK01012570 AJVK01012571 CRL05664.1 GGFK01004664 MBW37985.1 GGFJ01003394 MBW52535.1 GGFJ01003372 MBW52513.1 GGFM01001842 MBW22593.1 GGFL01002700 MBW66878.1 GEZM01051081 JAV75199.1 GGFL01002686 MBW66864.1 GGFJ01003279 MBW52420.1 GFDL01003963 JAV31082.1 GANP01004098 JAB80370.1 CH477974 EAT34689.1 CH477723 EAT36891.1 GBYB01011048 JAG80815.1 GDIQ01246721 JAK05004.1 KK853313 KDR08625.1 GFDF01008225 JAV05859.1 ATLV01019238 KE525266 KFB43966.1 GGFK01007626 MBW40947.1 GGFJ01003353 MBW52494.1 GBYB01011046 JAG80813.1 CRL05662.1 LJIJ01001104 ODM92975.1 GDIP01214029 JAJ09373.1 GDIP01143545 JAJ79857.1 GBYB01011044 JAG80811.1 GGFL01002681 MBW66859.1 GGFM01006850 MBW27601.1 AAAB01008968 EAA13295.5 KK117621 KFM70980.1 GGLE01005776 MBY09902.1 GFWV01022999 MAA47726.1 GFJQ02000470 JAW06500.1 GDIP01187030 JAJ36372.1 GDIP01189224 JAJ34178.1 GL732526 EFX88004.1 AJWK01029714 GDIP01170066 JAJ53336.1 KK113976 KFM61485.1 KB632375 ERL93890.1 AXCP01007737 GDIP01151823 JAJ71579.1 GANP01004097 JAB80371.1 GEFM01002764 JAP73032.1 GFAC01002641 JAT96547.1 GDIP01012925 JAM90790.1 GDIQ01015549 JAN79188.1
Proteomes
UP000053268
UP000005204
UP000007151
UP000218220
UP000053240
UP000079169
+ More
UP000007266 UP000192223 UP000009046 UP000005203 UP000076502 UP000001555 UP000235965 UP000183832 UP000002358 UP000069940 UP000249989 UP000092462 UP000075881 UP000075900 UP000008820 UP000027135 UP000030765 UP000075885 UP000094527 UP000007062 UP000054359 UP000000305 UP000092461 UP000030742 UP000075920 UP000075880
UP000007266 UP000192223 UP000009046 UP000005203 UP000076502 UP000001555 UP000235965 UP000183832 UP000002358 UP000069940 UP000249989 UP000092462 UP000075881 UP000075900 UP000008820 UP000027135 UP000030765 UP000075885 UP000094527 UP000007062 UP000054359 UP000000305 UP000092461 UP000030742 UP000075920 UP000075880
ProteinModelPortal
U6C886
A0A194Q2Q2
H9JRP2
A0A212EWG3
A0A2A4K8F4
U6C771
+ More
S4PMP0 A0A2A4K818 A0A0N1PHD8 A0A1B6DPS9 A0A3Q0IRJ0 A0A1B6E8X5 A0A0V0GCW0 A0A146LE46 V5G3G0 A0A1Y1LQU4 A0A1Y1LNI5 A0A1B6F6X4 A0A0A9WWL7 D6WBT5 A0A023F3G4 A0A1B6CIC4 A0A146L0J9 A0A1B6DFI6 A0A1W4XKX9 A0A1Y1LSE5 E0VPP1 T1IZ74 A0A224XQP1 A0A146LV70 A0A088ALZ2 A0A069DUT5 A0A0A9X6Z6 A0A154PK00 A0A2D3E2V8 A0A2R7WFT9 A0A310SES3 B7Q6T8 A0A2J7QQY3 A0A1J1IZU8 A0A0P4W827 K7JAU8 A0A2M4AAX5 A0A2M4CNX0 A0A182GN90 A0A2M4ABK6 A0A2M4AC57 A0A1B0D887 A0A1J1J010 A0A2M4AB23 A0A2M4BHK3 A0A182JSI6 A0A2M4BHP5 A0A182RJV7 A0A2M3Z261 A0A2M4CPR3 A0A1Y1LNJ8 A0A2M4CNH1 A0A2M4BH86 A0A1Q3FUE9 V5HAI1 Q16K77 Q16R65 A0A0C9Q9J5 A0A0P5G457 A0A067QIH5 A0A1L8DHC6 A0A084W172 A0A2M4AJM5 A0A182PMS5 A0A2M4BHJ7 A0A0C9RTY2 A0A1J1J2J0 A0A1D2MJD4 A0A0P4ZPX5 A0A0N8AIP9 A0A0C9R417 A0A2M4CNM3 A0A2M3ZGI4 Q7PMR9 A0A087U0U1 A0A2R5LK47 A0A293N4P5 A0A1Z5LGR6 A0A0P5BNK7 A0A0P5BIW9 E9FX36 A0A1B0CUW2 A0A0P5CII5 A0A087T8P6 U4UUQ9 A0A182WAX3 A0A182J471 A0A0P5DW26 V5IHZ1 A0A131Y3I2 A0A1E1XBB8 A0A0P6BQG8 A0A0P6HQS8
S4PMP0 A0A2A4K818 A0A0N1PHD8 A0A1B6DPS9 A0A3Q0IRJ0 A0A1B6E8X5 A0A0V0GCW0 A0A146LE46 V5G3G0 A0A1Y1LQU4 A0A1Y1LNI5 A0A1B6F6X4 A0A0A9WWL7 D6WBT5 A0A023F3G4 A0A1B6CIC4 A0A146L0J9 A0A1B6DFI6 A0A1W4XKX9 A0A1Y1LSE5 E0VPP1 T1IZ74 A0A224XQP1 A0A146LV70 A0A088ALZ2 A0A069DUT5 A0A0A9X6Z6 A0A154PK00 A0A2D3E2V8 A0A2R7WFT9 A0A310SES3 B7Q6T8 A0A2J7QQY3 A0A1J1IZU8 A0A0P4W827 K7JAU8 A0A2M4AAX5 A0A2M4CNX0 A0A182GN90 A0A2M4ABK6 A0A2M4AC57 A0A1B0D887 A0A1J1J010 A0A2M4AB23 A0A2M4BHK3 A0A182JSI6 A0A2M4BHP5 A0A182RJV7 A0A2M3Z261 A0A2M4CPR3 A0A1Y1LNJ8 A0A2M4CNH1 A0A2M4BH86 A0A1Q3FUE9 V5HAI1 Q16K77 Q16R65 A0A0C9Q9J5 A0A0P5G457 A0A067QIH5 A0A1L8DHC6 A0A084W172 A0A2M4AJM5 A0A182PMS5 A0A2M4BHJ7 A0A0C9RTY2 A0A1J1J2J0 A0A1D2MJD4 A0A0P4ZPX5 A0A0N8AIP9 A0A0C9R417 A0A2M4CNM3 A0A2M3ZGI4 Q7PMR9 A0A087U0U1 A0A2R5LK47 A0A293N4P5 A0A1Z5LGR6 A0A0P5BNK7 A0A0P5BIW9 E9FX36 A0A1B0CUW2 A0A0P5CII5 A0A087T8P6 U4UUQ9 A0A182WAX3 A0A182J471 A0A0P5DW26 V5IHZ1 A0A131Y3I2 A0A1E1XBB8 A0A0P6BQG8 A0A0P6HQS8
PDB
1RUT
E-value=2.27794e-11,
Score=166
Ontologies
PANTHER
Topology
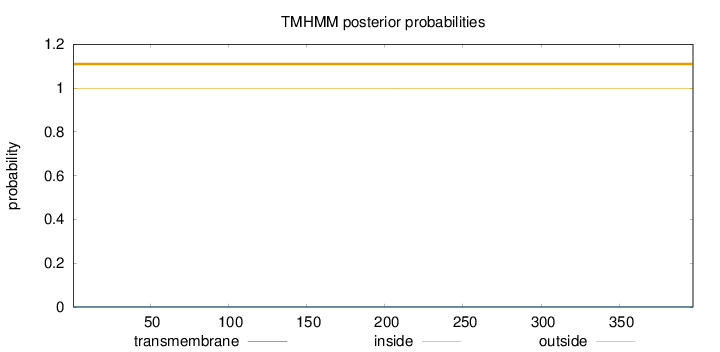
Length:
397
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000460000000000001
Exp number, first 60 AAs:
0.00031
Total prob of N-in:
0.00178
outside
1 - 397
Population Genetic Test Statistics
Pi
203.579302
Theta
201.604404
Tajima's D
0.417285
CLR
1.370448
CSRT
0.485575721213939
Interpretation
Uncertain
