Gene
KWMTBOMO14813
Pre Gene Modal
BGIBMGA012204
Annotation
RSM22-like_protein?_mitochondrial_[Operophtera_brumata]
Location in the cell
Mitochondrial Reliability : 2.34 Nuclear Reliability : 1.808
Sequence
CDS
ATGTTTCGTAAACTCAAATATTCACATATTTCGTCTTTAAGCGCCAACTACTCAACCAAAGTCTCAATAGATCCAAATTTAAAGGAAAGCTTTGACGCGCAGATCTATAAGCCACGGAAACATCCAGGAACCAATCGTGTAAAAATAGCATCAATACCTGCGGATATTCGGAAGGCAATTAAAATAATCCTAGACGATGCACGAGCTAAATCTTTACATCAGGAGAGCATTAAGCTCAAAAACCATCTGCAGTCTAGGCTACTGCCGCCAGAAGAAAAGGAAATTAATTTGAAAGCCGCGCAAATTCACGACAGCATTTCACAACGGACATTCTCTAAAATAAACAAAGAATTGAGCCAGGAGGAGTTGCAAAACATGAAAAAACAGGTACAGAATACTGTATTTAATATATTAAAAACGAACGTGTACCGTTGGGGTAATATATCGTACGATAAACCTACTAGTCTTCAATATTTGATGACCAGGGCTGCCCCGGAATACGCTGTCTTGGTTCGCGTATTAGATGAGATCAGAAGGGTTTTACCAGATTACAAGCCACGAAGCTTTTTTGACTTTGGTTCCGGCGTCGGAACCGGTACATGGGCACTGAACACATATTGGAAAGGAGAAATTTATGAATATTTTTGTGTCGATACATCGGCTCACATGCACGATTTGGCACGCTTAATTTTGTGTGAAGGCAAGGACAACGTCGAATTACCGTATAAATGTTATTACCAGCGTCAATTTCTACCTGCTTCCACAGATCTGAGGTACAATATAGTTTTGTCCGCGTACTCATTATTTGAACTACCCTCGATGAAATCAAGATTAGAAACAATACAAAAACTATGGAACAAGACCGAAGATTTCCTAATAATCATCGAACACGGAACAAATGCCGGATTCAGAGCTGTAAACGAAGCCAGAGAATTCATTTTGAATCTCCCAAAAGACAGCAGTAATAGAGGTTACGCATTCTCTCCGTGTCCAAACGACAGTGTTTGTCCAAGATACCTCGACCATCAAACTCCATGTAACTTCTTAATGAAGTACGAATCGCTAGGCTACCATTCAAAGTCTGAAGTTTTAGCAGATTTGTACACATATGTTATTTTAAGGAAAGGTGATCGACCAAATGACGATCCACAGTGGCCTAGGATTGTAAGGGCTCCGATTGTCAGGTCCGGACACACGATCTGTCGGATGTGTACAGCACAAGGAGAGTTAAAAGAAATTATATTTTCTAAAGGGAAGCACGATCAGACTATGTACAGATGTGCTAGGTCTGCTAATTGGGGAGATCTTTTACCTATAAAATAA
Protein
MFRKLKYSHISSLSANYSTKVSIDPNLKESFDAQIYKPRKHPGTNRVKIASIPADIRKAIKIILDDARAKSLHQESIKLKNHLQSRLLPPEEKEINLKAAQIHDSISQRTFSKINKELSQEELQNMKKQVQNTVFNILKTNVYRWGNISYDKPTSLQYLMTRAAPEYAVLVRVLDEIRRVLPDYKPRSFFDFGSGVGTGTWALNTYWKGEIYEYFCVDTSAHMHDLARLILCEGKDNVELPYKCYYQRQFLPASTDLRYNIVLSAYSLFELPSMKSRLETIQKLWNKTEDFLIIIEHGTNAGFRAVNEAREFILNLPKDSSNRGYAFSPCPNDSVCPRYLDHQTPCNFLMKYESLGYHSKSEVLADLYTYVILRKGDRPNDDPQWPRIVRAPIVRSGHTICRMCTAQGELKEIIFSKGKHDQTMYRCARSANWGDLLPIK
Summary
Uniprot
H9JRP4
A0A1E1WB91
A0A2W1BGH6
A0A2H1VP70
A0A0L7L006
A0A2A4KAK7
+ More
A0A0N1PH39 A0A194Q1S9 A0A1W4X7B9 A0A2J7QJV5 A0A067QP49 A0A182HCK1 A0A182GI75 N6UGG0 A0A139WKD9 A0A1S4F5R1 Q17ED0 A0A2P8Z603 A0A336MME2 A0A1B6D0H8 B0W2N5 A0A1Q3F4Z2 X1WIV5 A0A1B6KGD8 A0A088AG90 A0A1B0ABA6 A0A1B0C547 A0A1A9WHI7 A0A1A9YGN5 T1JKY4 A0A1Y1NKX5 A0A151X6N9 A0A1B0G0H3 A0A1J1IL91 A0A1I8PCM7 W8C6G2 A0A0P4VSF8 R4G459 A0A0J7L4U1 A0A195FDX9 A0A0L0CGS4 A0A1B0CI71 A0A158NT12 F4WY74 A0A3L8D9Q8 A0A026WX61 A0A151I3Z8 A0A1A9VE64 E9JB90 A0A293MTC1 A0A034VQJ4 A0A2H8TNB9 A0A195EHH4 A0A0A1WVI0 A0A0K8TXX6 A0A1Z5KWS7 A0A226F1H8 A0A0A9W281 A0A1D2NIC2 A0A069DUL8 A0A232F246 B4MX31 A0A1W4UF49 A0A195CDA5 A0A224XCX0 B4HWD6 A0A146KPW7 B4Q8A6 A0A0M4EH63 A0A0M3QTJ9 A0A3B0JU49 A0A0J9QZ60 B4M9F8 A0A1I8MQF1 A0A0C9QQX9 A0A023F790 B4JCG3 B3MPB9 Q29NF2 B4NYQ1 B4G7U4 A0A3Q0JJ30 A0A1I8PCX4 A0A0K8RLK2 A0A2R5L7K8 A0A131YAG9 Q9VL60 E2AZT0 B4KH31 A0A2L2YJ90 G3MS09 A0A2L2YH28 B3N8L2 A0A131XRG8 E9G9G8 W4Z116 A0A1E1XFU8 A0A087TYU0 A0A224Z466 A0A131Z0P3
A0A0N1PH39 A0A194Q1S9 A0A1W4X7B9 A0A2J7QJV5 A0A067QP49 A0A182HCK1 A0A182GI75 N6UGG0 A0A139WKD9 A0A1S4F5R1 Q17ED0 A0A2P8Z603 A0A336MME2 A0A1B6D0H8 B0W2N5 A0A1Q3F4Z2 X1WIV5 A0A1B6KGD8 A0A088AG90 A0A1B0ABA6 A0A1B0C547 A0A1A9WHI7 A0A1A9YGN5 T1JKY4 A0A1Y1NKX5 A0A151X6N9 A0A1B0G0H3 A0A1J1IL91 A0A1I8PCM7 W8C6G2 A0A0P4VSF8 R4G459 A0A0J7L4U1 A0A195FDX9 A0A0L0CGS4 A0A1B0CI71 A0A158NT12 F4WY74 A0A3L8D9Q8 A0A026WX61 A0A151I3Z8 A0A1A9VE64 E9JB90 A0A293MTC1 A0A034VQJ4 A0A2H8TNB9 A0A195EHH4 A0A0A1WVI0 A0A0K8TXX6 A0A1Z5KWS7 A0A226F1H8 A0A0A9W281 A0A1D2NIC2 A0A069DUL8 A0A232F246 B4MX31 A0A1W4UF49 A0A195CDA5 A0A224XCX0 B4HWD6 A0A146KPW7 B4Q8A6 A0A0M4EH63 A0A0M3QTJ9 A0A3B0JU49 A0A0J9QZ60 B4M9F8 A0A1I8MQF1 A0A0C9QQX9 A0A023F790 B4JCG3 B3MPB9 Q29NF2 B4NYQ1 B4G7U4 A0A3Q0JJ30 A0A1I8PCX4 A0A0K8RLK2 A0A2R5L7K8 A0A131YAG9 Q9VL60 E2AZT0 B4KH31 A0A2L2YJ90 G3MS09 A0A2L2YH28 B3N8L2 A0A131XRG8 E9G9G8 W4Z116 A0A1E1XFU8 A0A087TYU0 A0A224Z466 A0A131Z0P3
Pubmed
19121390
28756777
26227816
26354079
24845553
26483478
+ More
23537049 18362917 19820115 17510324 29403074 28004739 24495485 27129103 26108605 21347285 21719571 30249741 24508170 21282665 25348373 25830018 28528879 25401762 27289101 26334808 28648823 17994087 26823975 22936249 25315136 25474469 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20798317 26561354 22216098 28049606 21292972 28503490 28797301 26830274
23537049 18362917 19820115 17510324 29403074 28004739 24495485 27129103 26108605 21347285 21719571 30249741 24508170 21282665 25348373 25830018 28528879 25401762 27289101 26334808 28648823 17994087 26823975 22936249 25315136 25474469 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20798317 26561354 22216098 28049606 21292972 28503490 28797301 26830274
EMBL
BABH01031877
GDQN01006857
JAT84197.1
KZ150242
PZC71900.1
ODYU01003634
+ More
SOQ42631.1 JTDY01004098 KOB68604.1 NWSH01000015 PCG80823.1 KQ460396 KPJ15353.1 KQ459580 KPI99283.1 NEVH01013549 PNF28864.1 KK853117 KDR11155.1 JXUM01127565 KQ567205 KXJ69558.1 JXUM01065665 KQ562359 KXJ76071.1 APGK01026943 APGK01026944 APGK01026945 KB740648 KB632054 ENN79731.1 ERL88343.1 KQ971328 KYB28386.1 CH477283 EAT44846.1 PYGN01000180 PSN51932.1 UFQT01001740 SSX31604.1 GEDC01018165 JAS19133.1 DS231827 EDS29343.1 GFDL01012428 JAV22617.1 ABLF02009804 GEBQ01029492 JAT10485.1 JXJN01025826 JH431850 GEZM01003500 JAV96766.1 KQ982476 KYQ56061.1 CCAG010007529 CVRI01000055 CRL00928.1 GAMC01001132 JAC05424.1 GDKW01001726 JAI54869.1 ACPB03000223 GAHY01001630 JAA75880.1 LBMM01000722 KMQ97636.1 KQ981636 KYN38875.1 JRES01000418 KNC31436.1 AJWK01013066 ADTU01025470 GL888439 EGI60831.1 QOIP01000011 RLU17217.1 KK107078 EZA60433.1 KQ976472 KYM83978.1 GL770938 EFZ09912.1 GFWV01003134 GFWV01018621 MAA43349.1 GAKP01015119 JAC43833.1 GFXV01002963 MBW14768.1 KQ978957 KYN27309.1 GBXI01011874 JAD02418.1 GDHF01033178 JAI19136.1 GFJQ02007552 JAV99417.1 LNIX01000001 OXA63328.1 GBHO01044629 GBHO01044628 JAF98974.1 JAF98975.1 LJIJ01000037 ODN04716.1 GBGD01001383 JAC87506.1 NNAY01001263 OXU24569.1 CH963857 EDW76670.1 KQ978023 KYM98058.1 GFTR01006179 JAW10247.1 CH480818 EDW52331.1 GDHC01020065 JAP98563.1 CM000361 EDX04428.1 CP012523 ALC40217.1 ALC39043.1 OUUW01000010 SPP85634.1 CM002910 KMY89372.1 CH940654 EDW57834.2 GBYB01003067 JAG72834.1 GBBI01001605 JAC17107.1 CH916368 EDW03117.1 CH902620 EDV32238.1 CH379060 EAL33390.2 CM000157 EDW88715.1 CH479180 EDW28442.1 GADI01001838 JAA71970.1 GGLE01001324 MBY05450.1 GEFM01000314 JAP75482.1 AE014134 AY070900 AAF52836.1 AAL48522.1 AHN54301.1 GL444277 EFN61059.1 CH933807 EDW12242.1 IAAA01027933 LAA07440.1 JO844660 AEO36277.1 IAAA01027934 LAA07442.1 CH954177 EDV58435.1 GEFH01000305 JAP68276.1 GL732536 EFX83878.1 AAGJ04067069 GFAC01001038 JAT98150.1 KK117371 KFM70279.1 GFPF01009726 MAA20872.1 GEDV01004195 JAP84362.1
SOQ42631.1 JTDY01004098 KOB68604.1 NWSH01000015 PCG80823.1 KQ460396 KPJ15353.1 KQ459580 KPI99283.1 NEVH01013549 PNF28864.1 KK853117 KDR11155.1 JXUM01127565 KQ567205 KXJ69558.1 JXUM01065665 KQ562359 KXJ76071.1 APGK01026943 APGK01026944 APGK01026945 KB740648 KB632054 ENN79731.1 ERL88343.1 KQ971328 KYB28386.1 CH477283 EAT44846.1 PYGN01000180 PSN51932.1 UFQT01001740 SSX31604.1 GEDC01018165 JAS19133.1 DS231827 EDS29343.1 GFDL01012428 JAV22617.1 ABLF02009804 GEBQ01029492 JAT10485.1 JXJN01025826 JH431850 GEZM01003500 JAV96766.1 KQ982476 KYQ56061.1 CCAG010007529 CVRI01000055 CRL00928.1 GAMC01001132 JAC05424.1 GDKW01001726 JAI54869.1 ACPB03000223 GAHY01001630 JAA75880.1 LBMM01000722 KMQ97636.1 KQ981636 KYN38875.1 JRES01000418 KNC31436.1 AJWK01013066 ADTU01025470 GL888439 EGI60831.1 QOIP01000011 RLU17217.1 KK107078 EZA60433.1 KQ976472 KYM83978.1 GL770938 EFZ09912.1 GFWV01003134 GFWV01018621 MAA43349.1 GAKP01015119 JAC43833.1 GFXV01002963 MBW14768.1 KQ978957 KYN27309.1 GBXI01011874 JAD02418.1 GDHF01033178 JAI19136.1 GFJQ02007552 JAV99417.1 LNIX01000001 OXA63328.1 GBHO01044629 GBHO01044628 JAF98974.1 JAF98975.1 LJIJ01000037 ODN04716.1 GBGD01001383 JAC87506.1 NNAY01001263 OXU24569.1 CH963857 EDW76670.1 KQ978023 KYM98058.1 GFTR01006179 JAW10247.1 CH480818 EDW52331.1 GDHC01020065 JAP98563.1 CM000361 EDX04428.1 CP012523 ALC40217.1 ALC39043.1 OUUW01000010 SPP85634.1 CM002910 KMY89372.1 CH940654 EDW57834.2 GBYB01003067 JAG72834.1 GBBI01001605 JAC17107.1 CH916368 EDW03117.1 CH902620 EDV32238.1 CH379060 EAL33390.2 CM000157 EDW88715.1 CH479180 EDW28442.1 GADI01001838 JAA71970.1 GGLE01001324 MBY05450.1 GEFM01000314 JAP75482.1 AE014134 AY070900 AAF52836.1 AAL48522.1 AHN54301.1 GL444277 EFN61059.1 CH933807 EDW12242.1 IAAA01027933 LAA07440.1 JO844660 AEO36277.1 IAAA01027934 LAA07442.1 CH954177 EDV58435.1 GEFH01000305 JAP68276.1 GL732536 EFX83878.1 AAGJ04067069 GFAC01001038 JAT98150.1 KK117371 KFM70279.1 GFPF01009726 MAA20872.1 GEDV01004195 JAP84362.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000053240
UP000053268
UP000192223
+ More
UP000235965 UP000027135 UP000069940 UP000249989 UP000019118 UP000030742 UP000007266 UP000008820 UP000245037 UP000002320 UP000007819 UP000005203 UP000092445 UP000092460 UP000091820 UP000092443 UP000075809 UP000092444 UP000183832 UP000095300 UP000015103 UP000036403 UP000078541 UP000037069 UP000092461 UP000005205 UP000007755 UP000279307 UP000053097 UP000078540 UP000078200 UP000078492 UP000198287 UP000094527 UP000215335 UP000007798 UP000192221 UP000078542 UP000001292 UP000000304 UP000092553 UP000268350 UP000008792 UP000095301 UP000001070 UP000007801 UP000001819 UP000002282 UP000008744 UP000079169 UP000000803 UP000000311 UP000009192 UP000008711 UP000000305 UP000007110 UP000054359
UP000235965 UP000027135 UP000069940 UP000249989 UP000019118 UP000030742 UP000007266 UP000008820 UP000245037 UP000002320 UP000007819 UP000005203 UP000092445 UP000092460 UP000091820 UP000092443 UP000075809 UP000092444 UP000183832 UP000095300 UP000015103 UP000036403 UP000078541 UP000037069 UP000092461 UP000005205 UP000007755 UP000279307 UP000053097 UP000078540 UP000078200 UP000078492 UP000198287 UP000094527 UP000215335 UP000007798 UP000192221 UP000078542 UP000001292 UP000000304 UP000092553 UP000268350 UP000008792 UP000095301 UP000001070 UP000007801 UP000001819 UP000002282 UP000008744 UP000079169 UP000000803 UP000000311 UP000009192 UP000008711 UP000000305 UP000007110 UP000054359
Interpro
SUPFAM
SSF53335
SSF53335
ProteinModelPortal
H9JRP4
A0A1E1WB91
A0A2W1BGH6
A0A2H1VP70
A0A0L7L006
A0A2A4KAK7
+ More
A0A0N1PH39 A0A194Q1S9 A0A1W4X7B9 A0A2J7QJV5 A0A067QP49 A0A182HCK1 A0A182GI75 N6UGG0 A0A139WKD9 A0A1S4F5R1 Q17ED0 A0A2P8Z603 A0A336MME2 A0A1B6D0H8 B0W2N5 A0A1Q3F4Z2 X1WIV5 A0A1B6KGD8 A0A088AG90 A0A1B0ABA6 A0A1B0C547 A0A1A9WHI7 A0A1A9YGN5 T1JKY4 A0A1Y1NKX5 A0A151X6N9 A0A1B0G0H3 A0A1J1IL91 A0A1I8PCM7 W8C6G2 A0A0P4VSF8 R4G459 A0A0J7L4U1 A0A195FDX9 A0A0L0CGS4 A0A1B0CI71 A0A158NT12 F4WY74 A0A3L8D9Q8 A0A026WX61 A0A151I3Z8 A0A1A9VE64 E9JB90 A0A293MTC1 A0A034VQJ4 A0A2H8TNB9 A0A195EHH4 A0A0A1WVI0 A0A0K8TXX6 A0A1Z5KWS7 A0A226F1H8 A0A0A9W281 A0A1D2NIC2 A0A069DUL8 A0A232F246 B4MX31 A0A1W4UF49 A0A195CDA5 A0A224XCX0 B4HWD6 A0A146KPW7 B4Q8A6 A0A0M4EH63 A0A0M3QTJ9 A0A3B0JU49 A0A0J9QZ60 B4M9F8 A0A1I8MQF1 A0A0C9QQX9 A0A023F790 B4JCG3 B3MPB9 Q29NF2 B4NYQ1 B4G7U4 A0A3Q0JJ30 A0A1I8PCX4 A0A0K8RLK2 A0A2R5L7K8 A0A131YAG9 Q9VL60 E2AZT0 B4KH31 A0A2L2YJ90 G3MS09 A0A2L2YH28 B3N8L2 A0A131XRG8 E9G9G8 W4Z116 A0A1E1XFU8 A0A087TYU0 A0A224Z466 A0A131Z0P3
A0A0N1PH39 A0A194Q1S9 A0A1W4X7B9 A0A2J7QJV5 A0A067QP49 A0A182HCK1 A0A182GI75 N6UGG0 A0A139WKD9 A0A1S4F5R1 Q17ED0 A0A2P8Z603 A0A336MME2 A0A1B6D0H8 B0W2N5 A0A1Q3F4Z2 X1WIV5 A0A1B6KGD8 A0A088AG90 A0A1B0ABA6 A0A1B0C547 A0A1A9WHI7 A0A1A9YGN5 T1JKY4 A0A1Y1NKX5 A0A151X6N9 A0A1B0G0H3 A0A1J1IL91 A0A1I8PCM7 W8C6G2 A0A0P4VSF8 R4G459 A0A0J7L4U1 A0A195FDX9 A0A0L0CGS4 A0A1B0CI71 A0A158NT12 F4WY74 A0A3L8D9Q8 A0A026WX61 A0A151I3Z8 A0A1A9VE64 E9JB90 A0A293MTC1 A0A034VQJ4 A0A2H8TNB9 A0A195EHH4 A0A0A1WVI0 A0A0K8TXX6 A0A1Z5KWS7 A0A226F1H8 A0A0A9W281 A0A1D2NIC2 A0A069DUL8 A0A232F246 B4MX31 A0A1W4UF49 A0A195CDA5 A0A224XCX0 B4HWD6 A0A146KPW7 B4Q8A6 A0A0M4EH63 A0A0M3QTJ9 A0A3B0JU49 A0A0J9QZ60 B4M9F8 A0A1I8MQF1 A0A0C9QQX9 A0A023F790 B4JCG3 B3MPB9 Q29NF2 B4NYQ1 B4G7U4 A0A3Q0JJ30 A0A1I8PCX4 A0A0K8RLK2 A0A2R5L7K8 A0A131YAG9 Q9VL60 E2AZT0 B4KH31 A0A2L2YJ90 G3MS09 A0A2L2YH28 B3N8L2 A0A131XRG8 E9G9G8 W4Z116 A0A1E1XFU8 A0A087TYU0 A0A224Z466 A0A131Z0P3
PDB
3G5L
E-value=0.0178583,
Score=90
Ontologies
GO
Topology
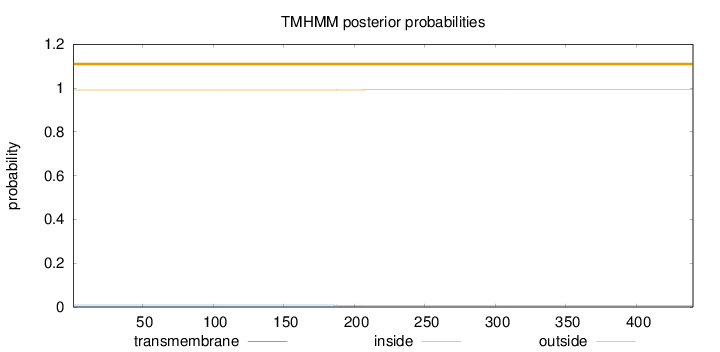
Length:
440
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01195
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00859
outside
1 - 440
Population Genetic Test Statistics
Pi
172.44418
Theta
174.002426
Tajima's D
-0.171468
CLR
371.960219
CSRT
0.328183590820459
Interpretation
Uncertain
