Gene
KWMTBOMO14805
Pre Gene Modal
BGIBMGA012139
Annotation
hypothetical_protein_KGM_01195_[Danaus_plexippus]
Location in the cell
Extracellular Reliability : 2.605
Sequence
CDS
ATGCAGAATATTCGCAAGATGATTACAATCTCCTTGAGCATCATGGTTCTACTCTGCAACGTGCCTTTGTCGCAAGCCATCGACTGTTTCAAATGCGTTTCAATGAACGGGAAATTCCCGCCATGCGACGATCCCTTCCACAACAATCATTCGCTGCAAATGCTGGAGGCGTCCTGTATGGGGGGCAGGAAGGGAAGGGACGGCCTGTTCCCTGCAACTTCGTGCATTAAAATAGCCGGTGTCTTCGATGACACAGGCGAAACCATAACAGTGCGAGGTTGTGGTTTAGACTCTGGTACGGCCACTACTGACACTGAAATCATAAGGATGTCTCATTGCGGAAGATTCTATTACAATAACAGATACGTCCATGGCTGTCTTCAGAGCTGCAACGACGCTGACGCCTGCAACTCGGCGAAGGAATATGGACCTTATGTGACAGTCTTGTCGCTCATTTACCTCGCGCTCATAATGTTCATGAGCTAA
Protein
MQNIRKMITISLSIMVLLCNVPLSQAIDCFKCVSMNGKFPPCDDPFHNNHSLQMLEASCMGGRKGRDGLFPATSCIKIAGVFDDTGETITVRGCGLDSGTATTDTEIIRMSHCGRFYYNNRYVHGCLQSCNDADACNSAKEYGPYVTVLSLIYLALIMFMS
Summary
Uniprot
H9JRH9
A0A2A4K8T6
A0A2H1VEU7
A0A0N1IN24
A0A0N0PCA6
A0A2W1B5T7
+ More
A0A182P054 A0A182JJA5 A0A182NRM1 A0A182RS65 A0A1B0FNK8 A0A1A9ZP44 A0A1A9X677 A0A1A9ULT7 A0A1B0DKN6 A0A182Q3V5 A0A1B0BM45 A0A182VE40 A0A182TM12 A0A182VWW3 A0A182M791 D6X4L9 A0A1I8MKI0 A0A182XAV6 A0A1S4GFF9 A0A1A9WM89 A0A182HSQ6 A0A182LES7 Q7QCC1 A0A182FNX5 B3LYN4 A0A182JWW5 Q17I18 W8BVR0 A0A336K312 A0A034WIW9 A0A1I8PI83 A0A182YSH9 A0A0A1WWH0 A0A1J1IHN0 B4QW35 Q8SYP1 B4ICK2 A0A0L0CB52 A0A182GPN9 B4PSV8 A0A1W4VKS0 A0A3B0JLT4 Q297A5 B4GEE5 B3P693 A0A212F460 B4NKU4 A0A1B0CKG2 B4LYU1 B4JGK6 A0A1B6G555 A0A084WSP8 B4K6E7 A0A1B6I1Q6 A0A088ATQ0 E2BJT7 A0A1B6MPE7 A0A1B6LX56 T1H7I1 A0A0L7QLT5 A0A1B6JUM9 F4WTN2 A0A151XB95 A0A158NG39 T1HNL2 A0A195E721 A0A195F1V6 E2AS25 A0A151I1R3 E9J5P7 A0A026X2J2 K7JE37 A0A0M4ERX4 A0A195CI24 A0A226F0Y8 E9GU67 A0A0N8C8S6 A0A1B6CUZ3 A0A1Y0F4B5 A0A0N8AHU7 A0A0P6GI44 A0A3L8DN52 A0A2S2Q2B3 C4WWG1 J9JJJ6 A0A2S2P560 A0A2S2P587 A0A2P6L889 A0A1S3D7B1
A0A182P054 A0A182JJA5 A0A182NRM1 A0A182RS65 A0A1B0FNK8 A0A1A9ZP44 A0A1A9X677 A0A1A9ULT7 A0A1B0DKN6 A0A182Q3V5 A0A1B0BM45 A0A182VE40 A0A182TM12 A0A182VWW3 A0A182M791 D6X4L9 A0A1I8MKI0 A0A182XAV6 A0A1S4GFF9 A0A1A9WM89 A0A182HSQ6 A0A182LES7 Q7QCC1 A0A182FNX5 B3LYN4 A0A182JWW5 Q17I18 W8BVR0 A0A336K312 A0A034WIW9 A0A1I8PI83 A0A182YSH9 A0A0A1WWH0 A0A1J1IHN0 B4QW35 Q8SYP1 B4ICK2 A0A0L0CB52 A0A182GPN9 B4PSV8 A0A1W4VKS0 A0A3B0JLT4 Q297A5 B4GEE5 B3P693 A0A212F460 B4NKU4 A0A1B0CKG2 B4LYU1 B4JGK6 A0A1B6G555 A0A084WSP8 B4K6E7 A0A1B6I1Q6 A0A088ATQ0 E2BJT7 A0A1B6MPE7 A0A1B6LX56 T1H7I1 A0A0L7QLT5 A0A1B6JUM9 F4WTN2 A0A151XB95 A0A158NG39 T1HNL2 A0A195E721 A0A195F1V6 E2AS25 A0A151I1R3 E9J5P7 A0A026X2J2 K7JE37 A0A0M4ERX4 A0A195CI24 A0A226F0Y8 E9GU67 A0A0N8C8S6 A0A1B6CUZ3 A0A1Y0F4B5 A0A0N8AHU7 A0A0P6GI44 A0A3L8DN52 A0A2S2Q2B3 C4WWG1 J9JJJ6 A0A2S2P560 A0A2S2P587 A0A2P6L889 A0A1S3D7B1
Pubmed
19121390
26354079
28756777
18362917
19820115
25315136
+ More
12364791 20966253 17994087 18057021 17510324 24495485 25348373 25244985 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605 26483478 17550304 15632085 22118469 24438588 20798317 21719571 21347285 21282665 24508170 20075255 21292972 30249741
12364791 20966253 17994087 18057021 17510324 24495485 25348373 25244985 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605 26483478 17550304 15632085 22118469 24438588 20798317 21719571 21347285 21282665 24508170 20075255 21292972 30249741
EMBL
BABH01031898
NWSH01000049
PCG80213.1
ODYU01002186
SOQ39355.1
KQ458863
+ More
KPJ04693.1 KQ460648 KPJ13144.1 KZ150302 PZC71529.1 CCAG010017775 AJVK01068041 AXCN02000004 JXJN01016665 JXJN01016666 AXCM01012478 KQ971380 EEZ97572.1 AAAB01008859 APCN01000320 EAA07696.4 CH902617 EDV42949.1 KPU80009.1 KPU80010.1 KPU80011.1 CH477243 EAT46341.1 GAMC01003259 GAMC01003258 GAMC01003257 JAC03298.1 UFQS01000097 UFQT01000097 SSW99384.1 SSX19764.1 GAKP01004862 JAC54090.1 GBXI01011301 JAD02991.1 CVRI01000050 CRK99266.1 CM000364 EDX14534.1 AY071413 AE014297 AAL49035.1 AAN14379.1 CH480828 EDW45098.1 JRES01000664 KNC29450.1 JXUM01015376 JXUM01015377 JXUM01015378 JXUM01015379 KQ560428 KXJ82461.1 CM000160 EDW98645.1 OUUW01000007 SPP83274.1 CM000070 EAL28301.1 CH479182 EDW33980.1 CH954182 EDV53563.1 AGBW02010436 OWR48519.1 CH964272 EDW84155.1 AJWK01016186 CH940650 EDW67018.1 KRF83056.1 KRF83057.1 KRF83058.1 KRF83059.1 CH916369 EDV93703.1 GECZ01012205 JAS57564.1 ATLV01026662 KE525416 KFB53242.1 CH933806 EDW15220.1 KRG01447.1 KRG01448.1 GECU01026843 JAS80863.1 GL448636 EFN84062.1 GEBQ01002147 JAT37830.1 GEBQ01011699 JAT28278.1 KQ414915 KOC59466.1 GECU01004796 JAT02911.1 GL888341 EGI62453.1 KQ982335 KYQ57651.1 ADTU01014953 ACPB03007662 KQ979568 KYN20995.1 KQ981864 KYN34152.1 GL442209 EFN63799.1 KQ976580 KYM79947.1 GL768174 EFZ11852.1 KK107020 EZA62525.1 CP012526 ALC45280.1 KQ977721 KYN00358.1 LNIX01000001 OXA63439.1 GL732565 EFX77025.1 GDIQ01103651 JAL48075.1 GEDC01019996 JAS17302.1 KX950847 ARU12057.1 GDIP01145961 JAJ77441.1 GDIQ01032885 JAN61852.1 QOIP01000006 RLU21801.1 GGMS01002714 MBY71917.1 AK342049 BAH72231.1 ABLF02028833 ABLF02028838 GGMR01011417 MBY24036.1 GGMR01011739 MBY24358.1 MWRG01001069 PRD34786.1
KPJ04693.1 KQ460648 KPJ13144.1 KZ150302 PZC71529.1 CCAG010017775 AJVK01068041 AXCN02000004 JXJN01016665 JXJN01016666 AXCM01012478 KQ971380 EEZ97572.1 AAAB01008859 APCN01000320 EAA07696.4 CH902617 EDV42949.1 KPU80009.1 KPU80010.1 KPU80011.1 CH477243 EAT46341.1 GAMC01003259 GAMC01003258 GAMC01003257 JAC03298.1 UFQS01000097 UFQT01000097 SSW99384.1 SSX19764.1 GAKP01004862 JAC54090.1 GBXI01011301 JAD02991.1 CVRI01000050 CRK99266.1 CM000364 EDX14534.1 AY071413 AE014297 AAL49035.1 AAN14379.1 CH480828 EDW45098.1 JRES01000664 KNC29450.1 JXUM01015376 JXUM01015377 JXUM01015378 JXUM01015379 KQ560428 KXJ82461.1 CM000160 EDW98645.1 OUUW01000007 SPP83274.1 CM000070 EAL28301.1 CH479182 EDW33980.1 CH954182 EDV53563.1 AGBW02010436 OWR48519.1 CH964272 EDW84155.1 AJWK01016186 CH940650 EDW67018.1 KRF83056.1 KRF83057.1 KRF83058.1 KRF83059.1 CH916369 EDV93703.1 GECZ01012205 JAS57564.1 ATLV01026662 KE525416 KFB53242.1 CH933806 EDW15220.1 KRG01447.1 KRG01448.1 GECU01026843 JAS80863.1 GL448636 EFN84062.1 GEBQ01002147 JAT37830.1 GEBQ01011699 JAT28278.1 KQ414915 KOC59466.1 GECU01004796 JAT02911.1 GL888341 EGI62453.1 KQ982335 KYQ57651.1 ADTU01014953 ACPB03007662 KQ979568 KYN20995.1 KQ981864 KYN34152.1 GL442209 EFN63799.1 KQ976580 KYM79947.1 GL768174 EFZ11852.1 KK107020 EZA62525.1 CP012526 ALC45280.1 KQ977721 KYN00358.1 LNIX01000001 OXA63439.1 GL732565 EFX77025.1 GDIQ01103651 JAL48075.1 GEDC01019996 JAS17302.1 KX950847 ARU12057.1 GDIP01145961 JAJ77441.1 GDIQ01032885 JAN61852.1 QOIP01000006 RLU21801.1 GGMS01002714 MBY71917.1 AK342049 BAH72231.1 ABLF02028833 ABLF02028838 GGMR01011417 MBY24036.1 GGMR01011739 MBY24358.1 MWRG01001069 PRD34786.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000075885
UP000075880
+ More
UP000075884 UP000075900 UP000092444 UP000092445 UP000092443 UP000078200 UP000092462 UP000075886 UP000092460 UP000075903 UP000075902 UP000075920 UP000075883 UP000007266 UP000095301 UP000076407 UP000091820 UP000075840 UP000075882 UP000007062 UP000069272 UP000007801 UP000075881 UP000008820 UP000095300 UP000076408 UP000183832 UP000000304 UP000000803 UP000001292 UP000037069 UP000069940 UP000249989 UP000002282 UP000192221 UP000268350 UP000001819 UP000008744 UP000008711 UP000007151 UP000007798 UP000092461 UP000008792 UP000001070 UP000030765 UP000009192 UP000005203 UP000008237 UP000015102 UP000053825 UP000007755 UP000075809 UP000005205 UP000015103 UP000078492 UP000078541 UP000000311 UP000078540 UP000053097 UP000002358 UP000092553 UP000078542 UP000198287 UP000000305 UP000279307 UP000007819 UP000079169
UP000075884 UP000075900 UP000092444 UP000092445 UP000092443 UP000078200 UP000092462 UP000075886 UP000092460 UP000075903 UP000075902 UP000075920 UP000075883 UP000007266 UP000095301 UP000076407 UP000091820 UP000075840 UP000075882 UP000007062 UP000069272 UP000007801 UP000075881 UP000008820 UP000095300 UP000076408 UP000183832 UP000000304 UP000000803 UP000001292 UP000037069 UP000069940 UP000249989 UP000002282 UP000192221 UP000268350 UP000001819 UP000008744 UP000008711 UP000007151 UP000007798 UP000092461 UP000008792 UP000001070 UP000030765 UP000009192 UP000005203 UP000008237 UP000015102 UP000053825 UP000007755 UP000075809 UP000005205 UP000015103 UP000078492 UP000078541 UP000000311 UP000078540 UP000053097 UP000002358 UP000092553 UP000078542 UP000198287 UP000000305 UP000279307 UP000007819 UP000079169
SUPFAM
SSF51905
SSF51905
Gene 3D
ProteinModelPortal
H9JRH9
A0A2A4K8T6
A0A2H1VEU7
A0A0N1IN24
A0A0N0PCA6
A0A2W1B5T7
+ More
A0A182P054 A0A182JJA5 A0A182NRM1 A0A182RS65 A0A1B0FNK8 A0A1A9ZP44 A0A1A9X677 A0A1A9ULT7 A0A1B0DKN6 A0A182Q3V5 A0A1B0BM45 A0A182VE40 A0A182TM12 A0A182VWW3 A0A182M791 D6X4L9 A0A1I8MKI0 A0A182XAV6 A0A1S4GFF9 A0A1A9WM89 A0A182HSQ6 A0A182LES7 Q7QCC1 A0A182FNX5 B3LYN4 A0A182JWW5 Q17I18 W8BVR0 A0A336K312 A0A034WIW9 A0A1I8PI83 A0A182YSH9 A0A0A1WWH0 A0A1J1IHN0 B4QW35 Q8SYP1 B4ICK2 A0A0L0CB52 A0A182GPN9 B4PSV8 A0A1W4VKS0 A0A3B0JLT4 Q297A5 B4GEE5 B3P693 A0A212F460 B4NKU4 A0A1B0CKG2 B4LYU1 B4JGK6 A0A1B6G555 A0A084WSP8 B4K6E7 A0A1B6I1Q6 A0A088ATQ0 E2BJT7 A0A1B6MPE7 A0A1B6LX56 T1H7I1 A0A0L7QLT5 A0A1B6JUM9 F4WTN2 A0A151XB95 A0A158NG39 T1HNL2 A0A195E721 A0A195F1V6 E2AS25 A0A151I1R3 E9J5P7 A0A026X2J2 K7JE37 A0A0M4ERX4 A0A195CI24 A0A226F0Y8 E9GU67 A0A0N8C8S6 A0A1B6CUZ3 A0A1Y0F4B5 A0A0N8AHU7 A0A0P6GI44 A0A3L8DN52 A0A2S2Q2B3 C4WWG1 J9JJJ6 A0A2S2P560 A0A2S2P587 A0A2P6L889 A0A1S3D7B1
A0A182P054 A0A182JJA5 A0A182NRM1 A0A182RS65 A0A1B0FNK8 A0A1A9ZP44 A0A1A9X677 A0A1A9ULT7 A0A1B0DKN6 A0A182Q3V5 A0A1B0BM45 A0A182VE40 A0A182TM12 A0A182VWW3 A0A182M791 D6X4L9 A0A1I8MKI0 A0A182XAV6 A0A1S4GFF9 A0A1A9WM89 A0A182HSQ6 A0A182LES7 Q7QCC1 A0A182FNX5 B3LYN4 A0A182JWW5 Q17I18 W8BVR0 A0A336K312 A0A034WIW9 A0A1I8PI83 A0A182YSH9 A0A0A1WWH0 A0A1J1IHN0 B4QW35 Q8SYP1 B4ICK2 A0A0L0CB52 A0A182GPN9 B4PSV8 A0A1W4VKS0 A0A3B0JLT4 Q297A5 B4GEE5 B3P693 A0A212F460 B4NKU4 A0A1B0CKG2 B4LYU1 B4JGK6 A0A1B6G555 A0A084WSP8 B4K6E7 A0A1B6I1Q6 A0A088ATQ0 E2BJT7 A0A1B6MPE7 A0A1B6LX56 T1H7I1 A0A0L7QLT5 A0A1B6JUM9 F4WTN2 A0A151XB95 A0A158NG39 T1HNL2 A0A195E721 A0A195F1V6 E2AS25 A0A151I1R3 E9J5P7 A0A026X2J2 K7JE37 A0A0M4ERX4 A0A195CI24 A0A226F0Y8 E9GU67 A0A0N8C8S6 A0A1B6CUZ3 A0A1Y0F4B5 A0A0N8AHU7 A0A0P6GI44 A0A3L8DN52 A0A2S2Q2B3 C4WWG1 J9JJJ6 A0A2S2P560 A0A2S2P587 A0A2P6L889 A0A1S3D7B1
Ontologies
Topology
SignalP
Position: 1 - 26,
Likelihood: 0.998107
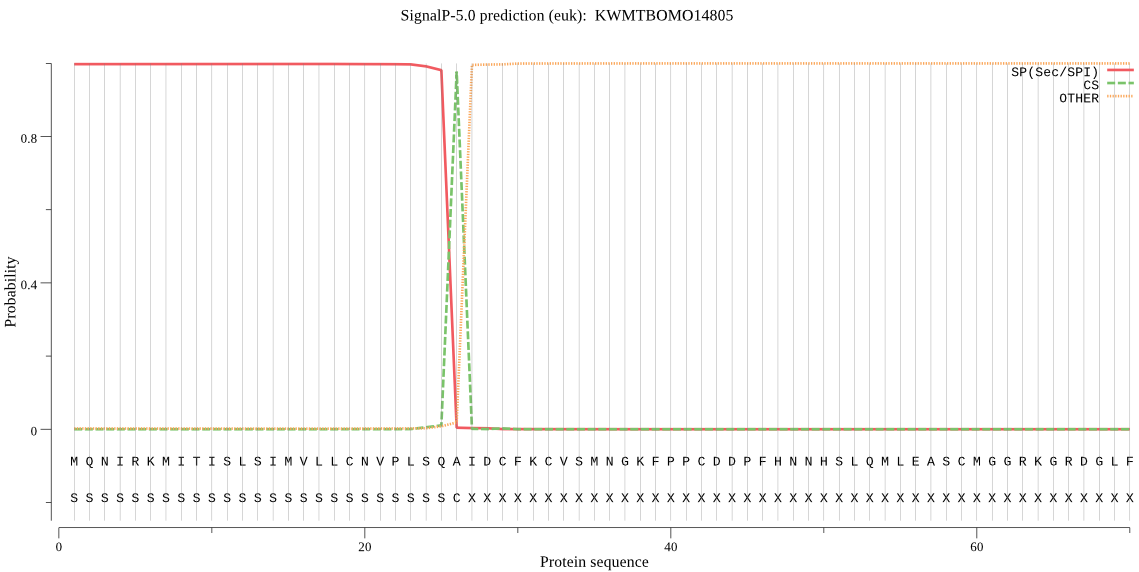
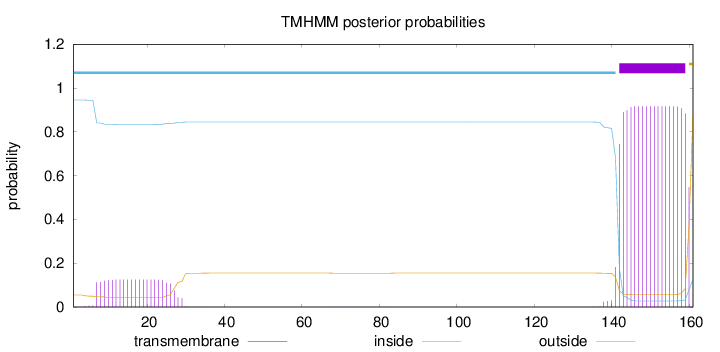
Length:
161
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
19.67574
Exp number, first 60 AAs:
2.61759
Total prob of N-in:
0.94492
inside
1 - 141
TMhelix
142 - 159
outside
160 - 161
Population Genetic Test Statistics
Pi
188.760057
Theta
2.868234
Tajima's D
1.614342
CLR
0.377339
CSRT
0.844457777111144
Interpretation
Uncertain
