Pre Gene Modal
BGIBMGA012211
Annotation
PREDICTED:_programmed_cell_death_protein_4-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 3.198
Sequence
CDS
ATGGAAGTCGATCGCATCGCTTCCGACTCTGAAAACGTGGCGATGGACGAAAAGCCGGTCGTAGAACAGGGCGCCGGAGACGCTGCGACGTCCAATCCCGAGCGATTACGTCGCAAGAACAAGAAATACGCTCGTTCTAACAGCAAGGACGGGCCGAGCGCTGTGACGCCGCTCCCGAAATATCGTAGCTGGAAGAACAGCCGAAGACCACGCAATGGCCACGGCAGGGGGTTACCGAAGAAAGGTGGGGCGGGAGGCAAAGGCGTGTGGGGTCTGCCCGGATCTGAGATGCTCGAGGAGTACGTGGAGGACCAGAACGATCCGAACTACGATTCGGAGGCGGTCACCAACGGCGACATCGAATTCAAGCAGGTCATAGTCGAAGCGGAGCCCGAGGATATTATCCGTAAATCCGAGCCTGTGATATTAGAGTATTTTGAGCATGGAGACACGAACGCGGCCGCCGAGGAGTTCCTAGACTTTGTGACGGCGGCGAGGAGTCATCTGGTGTGCGAGACAATTGTGGAGATAGCGTTGGATCACAAGGCGATGCACTGCGAAATGGCCTCGGTCCTCATATCCGATCTGTATGGAAGAGTTTTTTCAGCCAAAGACATCGGCTACGCGTTCGAGCGTCTGCTGGAGAAGCTGCCGGACCTGGTGCTGGACACGCCGGACGCGGCGCTGCTGCTGTCCAACTTCATAGCGCGCTGCGTGGCCGACGACTGCCTGCCGCCGCGCTTCGTGCAGGCGCAGGCCGCCGCCGCGCTCTCCGCGCCCGCCAGACAAGCTATACACAGAGCTGAGACTCTACTGTCTATGAAGCAAGGGCTAGTGAGGCTGGACAACATATGGGGCGTCGGCGGAGGTATCAGGCCGGTCAAGTCTCTCATCCGTCAGATCCAGTTGCTGCTGAAGGAGTACCTGACTTCGGGCGAGCTCGCGGAGGCGATGCGCTGCGTGCGGGAGTTGGAGGTTCCTCACTTCCACCACGAGCTGGTCTACGAGACAATACTCCTGGCTTTGGAGACCATAAATTCTGGTGTTGAAGAACAGCTTTGCACGTTTCTTGCTGAATTGCGGAGGTGCTGTATAGTCACGCCCGACCAAATGGACAGGGGCTTCATCCGCGTGCTCGAGGATATGAATGATATCGTCCTGGACGTGCCTCTCGCTTACATCATGCTGGACAGGTTCTTGGAGCGGTGCCAGACGAGGTTCAGGCTCGGGGACAACGTGCTGAAGCGTGTGCCGACGAGAGGTCGCAAGCGCTTCGTGTCCGAAGGAGACGGGGGCGCCATCAAGGATCACGCGCTGAAGCTGCGCGAGTGA
Protein
MEVDRIASDSENVAMDEKPVVEQGAGDAATSNPERLRRKNKKYARSNSKDGPSAVTPLPKYRSWKNSRRPRNGHGRGLPKKGGAGGKGVWGLPGSEMLEEYVEDQNDPNYDSEAVTNGDIEFKQVIVEAEPEDIIRKSEPVILEYFEHGDTNAAAEEFLDFVTAARSHLVCETIVEIALDHKAMHCEMASVLISDLYGRVFSAKDIGYAFERLLEKLPDLVLDTPDAALLLSNFIARCVADDCLPPRFVQAQAAAALSAPARQAIHRAETLLSMKQGLVRLDNIWGVGGGIRPVKSLIRQIQLLLKEYLTSGELAEAMRCVRELEVPHFHHELVYETILLALETINSGVEEQLCTFLAELRRCCIVTPDQMDRGFIRVLEDMNDIVLDVPLAYIMLDRFLERCQTRFRLGDNVLKRVPTRGRKRFVSEGDGGAIKDHALKLRE
Summary
Uniprot
A0A2A4K7K2
A0A1E1W475
S4PC15
A0A212FIK6
A0A212F461
A0A0N0PAF5
+ More
A0A2J7QSG0 A0A0L7L1Q6 A0A067QIK1 A0A0V0G3E2 A0A1W4WT55 A0A069DZ08 A0A224XGH1 A0A0T6B278 A0A0A9WWJ2 A0A1B6D254 A0A0K8TJ43 A0A0P4VUE5 R4FPP6 T1I1S7 A0A0K8TJP3 A0A034VEW0 A0A1L8DLU4 A0A1B6FUN2 A0A1Y1K444 A0A1B6LV66 A0A139WNU3 A0A1I8PVG0 A0A0K8VCS3 W8AY20 A0A0A1WK73 T1PCP1 A0A1I8MS75 D6WCT0 A0A0L0C0V7 D3TLD2 A0A023ESW1 U5ER32 A0A0K8TR71 A0A170Z2Q9 Q178N5 A0A1S4FBU7 A0A139WNA9 A0A182YCV3 A0A1Q3F416 A0A0A9WN83 N6T432 A0A0M4F8Y5 A0A1Q3F450 B4L2D8 A0A1Q3F435 A0A182MEH7 A0A1Q3F496 B4JL70 B4N1Z3 A0A1Q3F498 A0A0K8T7B4 B4M1I6 A0A3B0J7L6 A0A0P9AB19 B3MW12 A0A182QIS6 B0X887 B4Q2F7 A0A1W4VGL6 B5DM27 B4GY82 B3NVZ4 A0A182XJZ8 A0A1B0CHF0 A0A182VB79 A0A2M4BLG1 A0A1A9XZZ5 A0A1A9Z209 A0A336MNK8 A0A1A9VD60 A0A1A9WAQ5 A0A1B0B3Z7 W5JS62 Q9VY91 Q7QFA1 A0A1B0G2U4 E0VG43 A0A3R7NZL3 A0A182RHN7 A0A2M4A997 A0A1D2N058 A0A2M3Z1F2 A0A2M3Z1D0 A0A2M3Z1A1 V5I0L4 A0A182N8I4 L7MAM7 A0A182IPZ4 A0A023FIX2 A0A224Z6H7 A0A131YUA9
A0A2J7QSG0 A0A0L7L1Q6 A0A067QIK1 A0A0V0G3E2 A0A1W4WT55 A0A069DZ08 A0A224XGH1 A0A0T6B278 A0A0A9WWJ2 A0A1B6D254 A0A0K8TJ43 A0A0P4VUE5 R4FPP6 T1I1S7 A0A0K8TJP3 A0A034VEW0 A0A1L8DLU4 A0A1B6FUN2 A0A1Y1K444 A0A1B6LV66 A0A139WNU3 A0A1I8PVG0 A0A0K8VCS3 W8AY20 A0A0A1WK73 T1PCP1 A0A1I8MS75 D6WCT0 A0A0L0C0V7 D3TLD2 A0A023ESW1 U5ER32 A0A0K8TR71 A0A170Z2Q9 Q178N5 A0A1S4FBU7 A0A139WNA9 A0A182YCV3 A0A1Q3F416 A0A0A9WN83 N6T432 A0A0M4F8Y5 A0A1Q3F450 B4L2D8 A0A1Q3F435 A0A182MEH7 A0A1Q3F496 B4JL70 B4N1Z3 A0A1Q3F498 A0A0K8T7B4 B4M1I6 A0A3B0J7L6 A0A0P9AB19 B3MW12 A0A182QIS6 B0X887 B4Q2F7 A0A1W4VGL6 B5DM27 B4GY82 B3NVZ4 A0A182XJZ8 A0A1B0CHF0 A0A182VB79 A0A2M4BLG1 A0A1A9XZZ5 A0A1A9Z209 A0A336MNK8 A0A1A9VD60 A0A1A9WAQ5 A0A1B0B3Z7 W5JS62 Q9VY91 Q7QFA1 A0A1B0G2U4 E0VG43 A0A3R7NZL3 A0A182RHN7 A0A2M4A997 A0A1D2N058 A0A2M3Z1F2 A0A2M3Z1D0 A0A2M3Z1A1 V5I0L4 A0A182N8I4 L7MAM7 A0A182IPZ4 A0A023FIX2 A0A224Z6H7 A0A131YUA9
Pubmed
23622113
22118469
26354079
26227816
24845553
26334808
+ More
25401762 27129103 25348373 28004739 18362917 19820115 24495485 25830018 25315136 26108605 20353571 24945155 26369729 17510324 25244985 23537049 17994087 18057021 17550304 15632085 23185243 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12364791 20566863 27289101 25765539 25576852 28797301 26830274
25401762 27129103 25348373 28004739 18362917 19820115 24495485 25830018 25315136 26108605 20353571 24945155 26369729 17510324 25244985 23537049 17994087 18057021 17550304 15632085 23185243 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12364791 20566863 27289101 25765539 25576852 28797301 26830274
EMBL
NWSH01000049
PCG80211.1
GDQN01009266
JAT81788.1
GAIX01004356
JAA88204.1
+ More
AGBW02008362 OWR53570.1 AGBW02010436 OWR48520.1 KQ458863 KPJ04696.1 NEVH01011876 PNF31524.1 JTDY01003547 KOB69365.1 KK853313 KDR08650.1 GECL01003621 JAP02503.1 GBGD01001465 JAC87424.1 GFTR01006292 JAW10134.1 LJIG01016163 KRT81426.1 GBHO01034394 JAG09210.1 GEDC01017516 JAS19782.1 GBRD01000209 JAG65612.1 GDKW01000940 JAI55655.1 GAHY01000113 JAA77397.1 ACPB03016273 GBRD01000205 JAG65616.1 GAKP01018647 JAC40305.1 GFDF01006703 JAV07381.1 GECZ01015938 JAS53831.1 GEZM01097804 JAV54096.1 GEBQ01012406 JAT27571.1 KQ971311 KYB29481.1 GDHF01015662 GDHF01013713 JAI36652.1 JAI38601.1 GAMC01020816 GAMC01020815 GAMC01020814 GAMC01020813 GAMC01020812 JAB85741.1 GBXI01015377 GBXI01003097 JAC98914.1 JAD11195.1 KA646537 KA649545 AFP61166.1 EEZ99057.2 JRES01001065 KNC25876.1 EZ422234 ADD18510.1 GAPW01001215 JAC12383.1 GANO01002991 JAB56880.1 GDAI01000731 JAI16872.1 GEMB01002669 JAS00524.1 CH477361 EAT42655.1 KYB29480.1 GFDL01012733 JAV22312.1 GBHO01034395 JAG09209.1 APGK01053429 APGK01053430 APGK01053431 APGK01053432 KB741223 KB631623 ENN72393.1 ERL84761.1 CP012528 ALC48570.1 GFDL01012703 JAV22342.1 CH933810 EDW07799.1 GFDL01012748 JAV22297.1 AXCM01001073 GFDL01012661 JAV22384.1 CH916370 EDW00323.1 CH963925 EDW78382.2 GFDL01012660 JAV22385.1 GBRD01004515 JAG61306.1 CH940651 EDW65540.1 OUUW01000003 SPP78097.1 CH902625 KPU75458.1 EDV35157.1 KPU75457.1 AXCN02002231 DS232478 EDS42356.1 CM000162 EDX01618.1 CH379064 EDY72544.1 KRT06432.1 CH479197 EDW27738.1 CH954180 EDV47159.1 AJWK01012304 AJWK01012305 AJWK01012306 AJWK01012307 AJWK01012308 GGFJ01004744 MBW53885.1 UFQS01000843 UFQT01000749 UFQT01000843 SSX07225.1 SSX26939.1 SSX27568.1 JXJN01008132 ADMH02000256 ETN67227.1 AE014298 BT023835 AAF48312.2 AAZ86756.1 ABW09411.1 AGB95374.1 AGB95375.1 AAAB01008846 EAA06267.4 CCAG010022379 DS235131 EEB12349.1 QCYY01002318 ROT71268.1 GGFK01004052 MBW37373.1 LJIJ01000339 ODM98620.1 GGFM01001517 MBW22268.1 GGFM01001573 MBW22324.1 GGFM01001525 MBW22276.1 GANP01001876 JAB82592.1 GACK01004821 JAA60213.1 AXCP01007732 GBBK01002611 JAC21871.1 GFPF01010784 MAA21930.1 GEDV01006821 JAP81736.1
AGBW02008362 OWR53570.1 AGBW02010436 OWR48520.1 KQ458863 KPJ04696.1 NEVH01011876 PNF31524.1 JTDY01003547 KOB69365.1 KK853313 KDR08650.1 GECL01003621 JAP02503.1 GBGD01001465 JAC87424.1 GFTR01006292 JAW10134.1 LJIG01016163 KRT81426.1 GBHO01034394 JAG09210.1 GEDC01017516 JAS19782.1 GBRD01000209 JAG65612.1 GDKW01000940 JAI55655.1 GAHY01000113 JAA77397.1 ACPB03016273 GBRD01000205 JAG65616.1 GAKP01018647 JAC40305.1 GFDF01006703 JAV07381.1 GECZ01015938 JAS53831.1 GEZM01097804 JAV54096.1 GEBQ01012406 JAT27571.1 KQ971311 KYB29481.1 GDHF01015662 GDHF01013713 JAI36652.1 JAI38601.1 GAMC01020816 GAMC01020815 GAMC01020814 GAMC01020813 GAMC01020812 JAB85741.1 GBXI01015377 GBXI01003097 JAC98914.1 JAD11195.1 KA646537 KA649545 AFP61166.1 EEZ99057.2 JRES01001065 KNC25876.1 EZ422234 ADD18510.1 GAPW01001215 JAC12383.1 GANO01002991 JAB56880.1 GDAI01000731 JAI16872.1 GEMB01002669 JAS00524.1 CH477361 EAT42655.1 KYB29480.1 GFDL01012733 JAV22312.1 GBHO01034395 JAG09209.1 APGK01053429 APGK01053430 APGK01053431 APGK01053432 KB741223 KB631623 ENN72393.1 ERL84761.1 CP012528 ALC48570.1 GFDL01012703 JAV22342.1 CH933810 EDW07799.1 GFDL01012748 JAV22297.1 AXCM01001073 GFDL01012661 JAV22384.1 CH916370 EDW00323.1 CH963925 EDW78382.2 GFDL01012660 JAV22385.1 GBRD01004515 JAG61306.1 CH940651 EDW65540.1 OUUW01000003 SPP78097.1 CH902625 KPU75458.1 EDV35157.1 KPU75457.1 AXCN02002231 DS232478 EDS42356.1 CM000162 EDX01618.1 CH379064 EDY72544.1 KRT06432.1 CH479197 EDW27738.1 CH954180 EDV47159.1 AJWK01012304 AJWK01012305 AJWK01012306 AJWK01012307 AJWK01012308 GGFJ01004744 MBW53885.1 UFQS01000843 UFQT01000749 UFQT01000843 SSX07225.1 SSX26939.1 SSX27568.1 JXJN01008132 ADMH02000256 ETN67227.1 AE014298 BT023835 AAF48312.2 AAZ86756.1 ABW09411.1 AGB95374.1 AGB95375.1 AAAB01008846 EAA06267.4 CCAG010022379 DS235131 EEB12349.1 QCYY01002318 ROT71268.1 GGFK01004052 MBW37373.1 LJIJ01000339 ODM98620.1 GGFM01001517 MBW22268.1 GGFM01001573 MBW22324.1 GGFM01001525 MBW22276.1 GANP01001876 JAB82592.1 GACK01004821 JAA60213.1 AXCP01007732 GBBK01002611 JAC21871.1 GFPF01010784 MAA21930.1 GEDV01006821 JAP81736.1
Proteomes
UP000218220
UP000007151
UP000053268
UP000235965
UP000037510
UP000027135
+ More
UP000192223 UP000015103 UP000007266 UP000095300 UP000095301 UP000037069 UP000008820 UP000076408 UP000019118 UP000030742 UP000092553 UP000009192 UP000075883 UP000001070 UP000007798 UP000008792 UP000268350 UP000007801 UP000075886 UP000002320 UP000002282 UP000192221 UP000001819 UP000008744 UP000008711 UP000076407 UP000092461 UP000075903 UP000092443 UP000092445 UP000078200 UP000091820 UP000092460 UP000000673 UP000000803 UP000007062 UP000092444 UP000009046 UP000283509 UP000075900 UP000094527 UP000075884 UP000075880
UP000192223 UP000015103 UP000007266 UP000095300 UP000095301 UP000037069 UP000008820 UP000076408 UP000019118 UP000030742 UP000092553 UP000009192 UP000075883 UP000001070 UP000007798 UP000008792 UP000268350 UP000007801 UP000075886 UP000002320 UP000002282 UP000192221 UP000001819 UP000008744 UP000008711 UP000076407 UP000092461 UP000075903 UP000092443 UP000092445 UP000078200 UP000091820 UP000092460 UP000000673 UP000000803 UP000007062 UP000092444 UP000009046 UP000283509 UP000075900 UP000094527 UP000075884 UP000075880
PRIDE
Pfam
PF02847 MA3
Interpro
SUPFAM
SSF48371
SSF48371
Gene 3D
ProteinModelPortal
A0A2A4K7K2
A0A1E1W475
S4PC15
A0A212FIK6
A0A212F461
A0A0N0PAF5
+ More
A0A2J7QSG0 A0A0L7L1Q6 A0A067QIK1 A0A0V0G3E2 A0A1W4WT55 A0A069DZ08 A0A224XGH1 A0A0T6B278 A0A0A9WWJ2 A0A1B6D254 A0A0K8TJ43 A0A0P4VUE5 R4FPP6 T1I1S7 A0A0K8TJP3 A0A034VEW0 A0A1L8DLU4 A0A1B6FUN2 A0A1Y1K444 A0A1B6LV66 A0A139WNU3 A0A1I8PVG0 A0A0K8VCS3 W8AY20 A0A0A1WK73 T1PCP1 A0A1I8MS75 D6WCT0 A0A0L0C0V7 D3TLD2 A0A023ESW1 U5ER32 A0A0K8TR71 A0A170Z2Q9 Q178N5 A0A1S4FBU7 A0A139WNA9 A0A182YCV3 A0A1Q3F416 A0A0A9WN83 N6T432 A0A0M4F8Y5 A0A1Q3F450 B4L2D8 A0A1Q3F435 A0A182MEH7 A0A1Q3F496 B4JL70 B4N1Z3 A0A1Q3F498 A0A0K8T7B4 B4M1I6 A0A3B0J7L6 A0A0P9AB19 B3MW12 A0A182QIS6 B0X887 B4Q2F7 A0A1W4VGL6 B5DM27 B4GY82 B3NVZ4 A0A182XJZ8 A0A1B0CHF0 A0A182VB79 A0A2M4BLG1 A0A1A9XZZ5 A0A1A9Z209 A0A336MNK8 A0A1A9VD60 A0A1A9WAQ5 A0A1B0B3Z7 W5JS62 Q9VY91 Q7QFA1 A0A1B0G2U4 E0VG43 A0A3R7NZL3 A0A182RHN7 A0A2M4A997 A0A1D2N058 A0A2M3Z1F2 A0A2M3Z1D0 A0A2M3Z1A1 V5I0L4 A0A182N8I4 L7MAM7 A0A182IPZ4 A0A023FIX2 A0A224Z6H7 A0A131YUA9
A0A2J7QSG0 A0A0L7L1Q6 A0A067QIK1 A0A0V0G3E2 A0A1W4WT55 A0A069DZ08 A0A224XGH1 A0A0T6B278 A0A0A9WWJ2 A0A1B6D254 A0A0K8TJ43 A0A0P4VUE5 R4FPP6 T1I1S7 A0A0K8TJP3 A0A034VEW0 A0A1L8DLU4 A0A1B6FUN2 A0A1Y1K444 A0A1B6LV66 A0A139WNU3 A0A1I8PVG0 A0A0K8VCS3 W8AY20 A0A0A1WK73 T1PCP1 A0A1I8MS75 D6WCT0 A0A0L0C0V7 D3TLD2 A0A023ESW1 U5ER32 A0A0K8TR71 A0A170Z2Q9 Q178N5 A0A1S4FBU7 A0A139WNA9 A0A182YCV3 A0A1Q3F416 A0A0A9WN83 N6T432 A0A0M4F8Y5 A0A1Q3F450 B4L2D8 A0A1Q3F435 A0A182MEH7 A0A1Q3F496 B4JL70 B4N1Z3 A0A1Q3F498 A0A0K8T7B4 B4M1I6 A0A3B0J7L6 A0A0P9AB19 B3MW12 A0A182QIS6 B0X887 B4Q2F7 A0A1W4VGL6 B5DM27 B4GY82 B3NVZ4 A0A182XJZ8 A0A1B0CHF0 A0A182VB79 A0A2M4BLG1 A0A1A9XZZ5 A0A1A9Z209 A0A336MNK8 A0A1A9VD60 A0A1A9WAQ5 A0A1B0B3Z7 W5JS62 Q9VY91 Q7QFA1 A0A1B0G2U4 E0VG43 A0A3R7NZL3 A0A182RHN7 A0A2M4A997 A0A1D2N058 A0A2M3Z1F2 A0A2M3Z1D0 A0A2M3Z1A1 V5I0L4 A0A182N8I4 L7MAM7 A0A182IPZ4 A0A023FIX2 A0A224Z6H7 A0A131YUA9
PDB
3EIQ
E-value=6.77256e-69,
Score=663
Ontologies
KEGG
GO
PANTHER
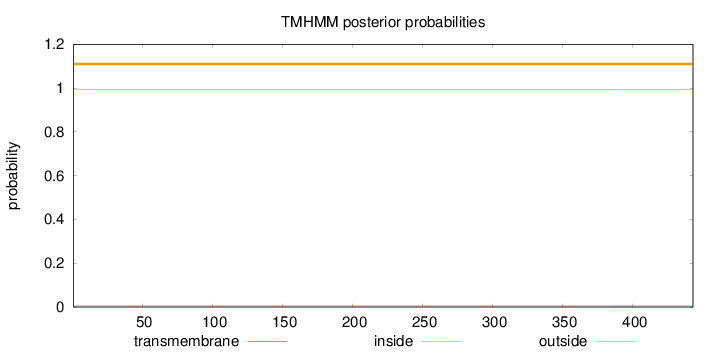
Topology
Length:
443
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00362
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00713
outside
1 - 443
Population Genetic Test Statistics
Pi
302.266582
Theta
149.89562
Tajima's D
0.633711
CLR
158.344282
CSRT
0.551422428878556
Interpretation
Uncertain
