Gene
KWMTBOMO14794
Pre Gene Modal
BGIBMGA012212
Annotation
PREDICTED:_charged_multivesicular_body_protein_7_[Bombyx_mori]
Transcription factor
Location in the cell
Cytoplasmic Reliability : 2.243
Sequence
CDS
ATGATGGGTCTACCCGACCGAGGAATACCCGAGGACAAGCTCCCGCAGTGTTGGTACGACGATGTCCGAATGAACGCACTCTTCGCCCCGTTTCGTTTAAAAACCACCAACCCTGAGTCCTGGGACATGAAGATGAAGTTCTGGTCTGATATGCTCAGGCAGTGGTGTAAGCACAGAAAGGACCCTATAGTGTCTCCTGCAGACGCAAGAGCAGCGTTCCAAAGAAAAGGACGCACTCCGGCCTGTCTTGATATTGTTATCGAAGAAATGTTTCGAAGCGGAGAGCTTTGTCCAATCTCAAAATATCAACGAATTCTTCACAACGGGACTGAGGGCTGGGTACGTTGGACAGCTCGACTTGCTCTCAAACCTGCTGCACTCGCTCTCACAGCTGTCTCTTCATTTCTACCTGCACGACAAACTCTGGACAATGATGGCTTGCCTAAAGCTAGTATAGAATGCACACAGAGGTTCCTACTTGAAGGATCGGTCAAGGAGCTAGCAACAGAACTACTTCACACAAACGTACCTGAAATAGACCGAATCGGTACTATAGAAGAATTAATGAAGAACTTTGAGTATCAAGGTGGCAGAGAAGTCTTCGAAGTGCTGCTCGGTTGGCTGGTAGCTCAGGGCTATGCAGTAAAAAAAGATGAAGTTATTAAACTGGCTGAACCAGATAAGAAAGCCACCCCCGTGACAGAGTCAGATGAAGCGCTTGTGAAGCTAGTCACAGCTGAGAGGCACTTGCACGGAGACTCCACAAGACTGTCCCGAGAACTAGCAACTATTGAGAATGATGCTCGAGCTGCACTTCAGCTAGGTAATAGGATTGCGGCCAAGAATCATCTGAGACGTAAGCACAAAGTGCAACGGAAACTGGAACAGTGCACGAAAGCTCTGGAAAACGTACAGAACTTACTACAACAGATGAGGGATGTACATTTCAATGCATCAATTGTCGATACTTACAAGACAACATCAGAAGCTATGAAGCGAGGATTAAAAGACAACGGTCTTGAGGTGGATACAGTTCACGACACGATGGACGATCTGAAAGAGGTAATGGATTCATATAGCGAAATGGAGAGAGCACTTGGTACAACAATAGACGACACCGACACAGCTGAGCTTGAAAGAGAATTGAAGGAACTGCTGTCGGGACCGTCGGCGCCTGGTGGCGGAACTCCCGGGAAGGAGGGGGAGGTGAAGGAATAA
Protein
MMGLPDRGIPEDKLPQCWYDDVRMNALFAPFRLKTTNPESWDMKMKFWSDMLRQWCKHRKDPIVSPADARAAFQRKGRTPACLDIVIEEMFRSGELCPISKYQRILHNGTEGWVRWTARLALKPAALALTAVSSFLPARQTLDNDGLPKASIECTQRFLLEGSVKELATELLHTNVPEIDRIGTIEELMKNFEYQGGREVFEVLLGWLVAQGYAVKKDEVIKLAEPDKKATPVTESDEALVKLVTAERHLHGDSTRLSRELATIENDARAALQLGNRIAAKNHLRRKHKVQRKLEQCTKALENVQNLLQQMRDVHFNASIVDTYKTTSEAMKRGLKDNGLEVDTVHDTMDDLKEVMDSYSEMERALGTTIDDTDTAELERELKELLSGPSAPGGGTPGKEGEVKE
Summary
Uniprot
A0A3S2NFQ0
A0A2A4JL95
A0A2W1BJP7
A0A194Q1P2
A0A2H1V192
A0A194R500
+ More
A0A0V0GAA9 A0A2L2Y6A8 K7IPF6 A0A1B6CL40 A0A088AS34 A0A232EKP0 A0A0M9A5D6 A0A069DT01 A0A023F7L9 A0A224XPV1 A0A151J6B3 A0A2A3E849 B0WFW4 A0A1Q3F7D4 A0A067QQ38 A0A1S3D855 F4W9T3 A0A195CFJ8 A0A158NYV4 A0A195EZ66 A0A310S7L8 A0A026WCP7 A0A195BS09 A0A151WKH0 A0A182GRB1 A0A1W4X132 T1J0Y3 A0A0L7RG44 A0A182INU5 A0A2J7RPT7 T1HR31 E2A5S8 R4WEC1 E2B4A8 A0A0P4VPN4 R4G811 A0A2P6KX35 A0A084W184 A0A182FT74 A0A182SNA2 A0A182RJU3 A0A182MBU3 A0A0L0CPZ8 Q16F51 Q16P72 A0A182NQB8 A0A1Y1KAX8 A0A1B6F3N6 J9JTF6 A0A2S2NB11 A0A2M4AJG2 U4UUL4 A0A1I8NDQ1 T1PHM1 A0A182WAY9 W8C2U3 J3JWH1 A0A1B0GIC9 N6U305 D6WCG1 A0A131XUA9 V5H4X7 A0A1A9YRR6 A0A2M4CIQ5 L7M7J4 A0A1B0B491 A0A0T6B2C5 A0A1B6I0D0 B7PN70 A0A1A9UF24 A0A023GJY9 G3MT65 A0A0A9Y3W9 A0A224YUD4 A0A131YYZ9 A0A2P1L4E0 A0A131XH18 A0A2M4CJZ3 A0A1L8DAL2 E9I970 A0A0P6DXD5 A0A1E1XDP8 A0A182QIJ9 A0A293LRU6 A0A0K0DIC0 A0A0A1WZX0 A0A2M4BQC7 A0A0M3KA03 W4Y2H9 F1L6Y9 A0A2G8L8I7
A0A0V0GAA9 A0A2L2Y6A8 K7IPF6 A0A1B6CL40 A0A088AS34 A0A232EKP0 A0A0M9A5D6 A0A069DT01 A0A023F7L9 A0A224XPV1 A0A151J6B3 A0A2A3E849 B0WFW4 A0A1Q3F7D4 A0A067QQ38 A0A1S3D855 F4W9T3 A0A195CFJ8 A0A158NYV4 A0A195EZ66 A0A310S7L8 A0A026WCP7 A0A195BS09 A0A151WKH0 A0A182GRB1 A0A1W4X132 T1J0Y3 A0A0L7RG44 A0A182INU5 A0A2J7RPT7 T1HR31 E2A5S8 R4WEC1 E2B4A8 A0A0P4VPN4 R4G811 A0A2P6KX35 A0A084W184 A0A182FT74 A0A182SNA2 A0A182RJU3 A0A182MBU3 A0A0L0CPZ8 Q16F51 Q16P72 A0A182NQB8 A0A1Y1KAX8 A0A1B6F3N6 J9JTF6 A0A2S2NB11 A0A2M4AJG2 U4UUL4 A0A1I8NDQ1 T1PHM1 A0A182WAY9 W8C2U3 J3JWH1 A0A1B0GIC9 N6U305 D6WCG1 A0A131XUA9 V5H4X7 A0A1A9YRR6 A0A2M4CIQ5 L7M7J4 A0A1B0B491 A0A0T6B2C5 A0A1B6I0D0 B7PN70 A0A1A9UF24 A0A023GJY9 G3MT65 A0A0A9Y3W9 A0A224YUD4 A0A131YYZ9 A0A2P1L4E0 A0A131XH18 A0A2M4CJZ3 A0A1L8DAL2 E9I970 A0A0P6DXD5 A0A1E1XDP8 A0A182QIJ9 A0A293LRU6 A0A0K0DIC0 A0A0A1WZX0 A0A2M4BQC7 A0A0M3KA03 W4Y2H9 F1L6Y9 A0A2G8L8I7
Pubmed
28756777
26354079
26561354
20075255
28648823
26334808
+ More
25474469 24845553 21719571 21347285 24508170 30249741 26483478 20798317 23691247 27129103 24438588 26108605 17510324 28004739 23537049 25315136 24495485 22516182 18362917 19820115 25576852 22216098 25401762 26823975 28797301 26830274 29501422 28049606 21282665 28503490 25830018 21685128 29023486
25474469 24845553 21719571 21347285 24508170 30249741 26483478 20798317 23691247 27129103 24438588 26108605 17510324 28004739 23537049 25315136 24495485 22516182 18362917 19820115 25576852 22216098 25401762 26823975 28797301 26830274 29501422 28049606 21282665 28503490 25830018 21685128 29023486
EMBL
RSAL01000132
RVE46375.1
NWSH01001067
PCG72827.1
KZ150102
PZC73487.1
+ More
KQ459580 KPI99243.1 ODYU01000216 SOQ34615.1 KQ460685 KPJ12757.1 GECL01001131 JAP04993.1 IAAA01017265 LAA03712.1 GEDC01023114 GEDC01019449 JAS14184.1 JAS17849.1 NNAY01003732 OXU18930.1 KQ435750 KOX76283.1 GBGD01001666 JAC87223.1 GBBI01001513 JAC17199.1 GFTR01005834 JAW10592.1 KQ979868 KYN18745.1 KZ288329 PBC27927.1 DS231921 EDS26499.1 GFDL01011569 JAV23476.1 KK853149 KDR10675.1 GL888033 EGI69069.1 KQ977876 KYM99091.1 ADTU01000801 KQ981909 KYN33169.1 KQ765323 KQ759877 OAD53989.1 OAD62272.1 KK107315 QOIP01000002 EZA52824.1 RLU25894.1 KQ976423 KYM88792.1 KQ983012 KYQ48336.1 JXUM01016395 KQ560457 KXJ82319.1 JH431742 KQ414599 KOC69794.1 NEVH01001355 PNF42844.1 ACPB03022348 GL437059 EFN71200.1 AK418446 BAN21608.1 GL445549 EFN89456.1 GDKW01002141 JAI54454.1 GAHY01001782 JAA75728.1 MWRG01004044 PRD30890.1 ATLV01019246 KE525266 KFB43978.1 AXCM01004171 JRES01000065 KNC34430.1 CH478510 EAT32866.1 CH477792 EAT36162.1 GEZM01087675 JAV58544.1 GECZ01024873 JAS44896.1 ABLF02032620 GGMR01001775 MBY14394.1 GGFK01007602 MBW40923.1 KB632375 ERL93876.1 KA648266 AFP62895.1 GAMC01006389 JAC00167.1 BT127589 AEE62551.1 AJWK01013691 APGK01051693 APGK01051694 KB741201 ENN72952.1 KQ971311 EEZ99020.1 GEFM01005559 JAP70237.1 GALX01000603 JAB67863.1 GGFL01001039 MBW65217.1 GACK01004763 JAA60271.1 JXJN01008218 JXJN01008219 JXJN01008220 LJIG01016130 KRT81526.1 GECU01027333 JAS80373.1 ABJB010214903 DS750732 EEC08042.1 GBBM01001179 JAC34239.1 JO845066 AEO36683.1 GBHO01019394 GBHO01019393 GBRD01011576 GDHC01017663 JAG24210.1 JAG24211.1 JAG54248.1 JAQ00966.1 GFPF01006727 MAA17873.1 GEDV01004775 JAP83782.1 MG596909 AVP12664.1 GEFH01003570 JAP65011.1 GGFL01001040 MBW65218.1 GFDF01010677 JAV03407.1 GL761745 EFZ22882.1 GDIQ01070910 LRGB01001005 JAN23827.1 KZS14083.1 GFAC01001952 JAT97236.1 AXCN02001893 GFWV01004725 MAA29455.1 GBXI01010354 JAD03938.1 GGFJ01006032 MBW55173.1 UYRR01033854 VDK59768.1 AAGJ04077408 JI172722 ADY45893.1 MRZV01000170 PIK56578.1
KQ459580 KPI99243.1 ODYU01000216 SOQ34615.1 KQ460685 KPJ12757.1 GECL01001131 JAP04993.1 IAAA01017265 LAA03712.1 GEDC01023114 GEDC01019449 JAS14184.1 JAS17849.1 NNAY01003732 OXU18930.1 KQ435750 KOX76283.1 GBGD01001666 JAC87223.1 GBBI01001513 JAC17199.1 GFTR01005834 JAW10592.1 KQ979868 KYN18745.1 KZ288329 PBC27927.1 DS231921 EDS26499.1 GFDL01011569 JAV23476.1 KK853149 KDR10675.1 GL888033 EGI69069.1 KQ977876 KYM99091.1 ADTU01000801 KQ981909 KYN33169.1 KQ765323 KQ759877 OAD53989.1 OAD62272.1 KK107315 QOIP01000002 EZA52824.1 RLU25894.1 KQ976423 KYM88792.1 KQ983012 KYQ48336.1 JXUM01016395 KQ560457 KXJ82319.1 JH431742 KQ414599 KOC69794.1 NEVH01001355 PNF42844.1 ACPB03022348 GL437059 EFN71200.1 AK418446 BAN21608.1 GL445549 EFN89456.1 GDKW01002141 JAI54454.1 GAHY01001782 JAA75728.1 MWRG01004044 PRD30890.1 ATLV01019246 KE525266 KFB43978.1 AXCM01004171 JRES01000065 KNC34430.1 CH478510 EAT32866.1 CH477792 EAT36162.1 GEZM01087675 JAV58544.1 GECZ01024873 JAS44896.1 ABLF02032620 GGMR01001775 MBY14394.1 GGFK01007602 MBW40923.1 KB632375 ERL93876.1 KA648266 AFP62895.1 GAMC01006389 JAC00167.1 BT127589 AEE62551.1 AJWK01013691 APGK01051693 APGK01051694 KB741201 ENN72952.1 KQ971311 EEZ99020.1 GEFM01005559 JAP70237.1 GALX01000603 JAB67863.1 GGFL01001039 MBW65217.1 GACK01004763 JAA60271.1 JXJN01008218 JXJN01008219 JXJN01008220 LJIG01016130 KRT81526.1 GECU01027333 JAS80373.1 ABJB010214903 DS750732 EEC08042.1 GBBM01001179 JAC34239.1 JO845066 AEO36683.1 GBHO01019394 GBHO01019393 GBRD01011576 GDHC01017663 JAG24210.1 JAG24211.1 JAG54248.1 JAQ00966.1 GFPF01006727 MAA17873.1 GEDV01004775 JAP83782.1 MG596909 AVP12664.1 GEFH01003570 JAP65011.1 GGFL01001040 MBW65218.1 GFDF01010677 JAV03407.1 GL761745 EFZ22882.1 GDIQ01070910 LRGB01001005 JAN23827.1 KZS14083.1 GFAC01001952 JAT97236.1 AXCN02001893 GFWV01004725 MAA29455.1 GBXI01010354 JAD03938.1 GGFJ01006032 MBW55173.1 UYRR01033854 VDK59768.1 AAGJ04077408 JI172722 ADY45893.1 MRZV01000170 PIK56578.1
Proteomes
UP000283053
UP000218220
UP000053268
UP000053240
UP000002358
UP000005203
+ More
UP000215335 UP000053105 UP000078492 UP000242457 UP000002320 UP000027135 UP000079169 UP000007755 UP000078542 UP000005205 UP000078541 UP000053097 UP000279307 UP000078540 UP000075809 UP000069940 UP000249989 UP000192223 UP000053825 UP000075880 UP000235965 UP000015103 UP000000311 UP000008237 UP000030765 UP000069272 UP000075901 UP000075900 UP000075883 UP000037069 UP000008820 UP000075884 UP000007819 UP000030742 UP000095301 UP000075920 UP000092461 UP000019118 UP000007266 UP000092443 UP000092460 UP000001555 UP000078200 UP000076858 UP000075886 UP000035642 UP000036680 UP000007110 UP000230750
UP000215335 UP000053105 UP000078492 UP000242457 UP000002320 UP000027135 UP000079169 UP000007755 UP000078542 UP000005205 UP000078541 UP000053097 UP000279307 UP000078540 UP000075809 UP000069940 UP000249989 UP000192223 UP000053825 UP000075880 UP000235965 UP000015103 UP000000311 UP000008237 UP000030765 UP000069272 UP000075901 UP000075900 UP000075883 UP000037069 UP000008820 UP000075884 UP000007819 UP000030742 UP000095301 UP000075920 UP000092461 UP000019118 UP000007266 UP000092443 UP000092460 UP000001555 UP000078200 UP000076858 UP000075886 UP000035642 UP000036680 UP000007110 UP000230750
Pfam
PF03357 Snf7
Interpro
IPR005024
Snf7_fam
ProteinModelPortal
A0A3S2NFQ0
A0A2A4JL95
A0A2W1BJP7
A0A194Q1P2
A0A2H1V192
A0A194R500
+ More
A0A0V0GAA9 A0A2L2Y6A8 K7IPF6 A0A1B6CL40 A0A088AS34 A0A232EKP0 A0A0M9A5D6 A0A069DT01 A0A023F7L9 A0A224XPV1 A0A151J6B3 A0A2A3E849 B0WFW4 A0A1Q3F7D4 A0A067QQ38 A0A1S3D855 F4W9T3 A0A195CFJ8 A0A158NYV4 A0A195EZ66 A0A310S7L8 A0A026WCP7 A0A195BS09 A0A151WKH0 A0A182GRB1 A0A1W4X132 T1J0Y3 A0A0L7RG44 A0A182INU5 A0A2J7RPT7 T1HR31 E2A5S8 R4WEC1 E2B4A8 A0A0P4VPN4 R4G811 A0A2P6KX35 A0A084W184 A0A182FT74 A0A182SNA2 A0A182RJU3 A0A182MBU3 A0A0L0CPZ8 Q16F51 Q16P72 A0A182NQB8 A0A1Y1KAX8 A0A1B6F3N6 J9JTF6 A0A2S2NB11 A0A2M4AJG2 U4UUL4 A0A1I8NDQ1 T1PHM1 A0A182WAY9 W8C2U3 J3JWH1 A0A1B0GIC9 N6U305 D6WCG1 A0A131XUA9 V5H4X7 A0A1A9YRR6 A0A2M4CIQ5 L7M7J4 A0A1B0B491 A0A0T6B2C5 A0A1B6I0D0 B7PN70 A0A1A9UF24 A0A023GJY9 G3MT65 A0A0A9Y3W9 A0A224YUD4 A0A131YYZ9 A0A2P1L4E0 A0A131XH18 A0A2M4CJZ3 A0A1L8DAL2 E9I970 A0A0P6DXD5 A0A1E1XDP8 A0A182QIJ9 A0A293LRU6 A0A0K0DIC0 A0A0A1WZX0 A0A2M4BQC7 A0A0M3KA03 W4Y2H9 F1L6Y9 A0A2G8L8I7
A0A0V0GAA9 A0A2L2Y6A8 K7IPF6 A0A1B6CL40 A0A088AS34 A0A232EKP0 A0A0M9A5D6 A0A069DT01 A0A023F7L9 A0A224XPV1 A0A151J6B3 A0A2A3E849 B0WFW4 A0A1Q3F7D4 A0A067QQ38 A0A1S3D855 F4W9T3 A0A195CFJ8 A0A158NYV4 A0A195EZ66 A0A310S7L8 A0A026WCP7 A0A195BS09 A0A151WKH0 A0A182GRB1 A0A1W4X132 T1J0Y3 A0A0L7RG44 A0A182INU5 A0A2J7RPT7 T1HR31 E2A5S8 R4WEC1 E2B4A8 A0A0P4VPN4 R4G811 A0A2P6KX35 A0A084W184 A0A182FT74 A0A182SNA2 A0A182RJU3 A0A182MBU3 A0A0L0CPZ8 Q16F51 Q16P72 A0A182NQB8 A0A1Y1KAX8 A0A1B6F3N6 J9JTF6 A0A2S2NB11 A0A2M4AJG2 U4UUL4 A0A1I8NDQ1 T1PHM1 A0A182WAY9 W8C2U3 J3JWH1 A0A1B0GIC9 N6U305 D6WCG1 A0A131XUA9 V5H4X7 A0A1A9YRR6 A0A2M4CIQ5 L7M7J4 A0A1B0B491 A0A0T6B2C5 A0A1B6I0D0 B7PN70 A0A1A9UF24 A0A023GJY9 G3MT65 A0A0A9Y3W9 A0A224YUD4 A0A131YYZ9 A0A2P1L4E0 A0A131XH18 A0A2M4CJZ3 A0A1L8DAL2 E9I970 A0A0P6DXD5 A0A1E1XDP8 A0A182QIJ9 A0A293LRU6 A0A0K0DIC0 A0A0A1WZX0 A0A2M4BQC7 A0A0M3KA03 W4Y2H9 F1L6Y9 A0A2G8L8I7
PDB
5FD7
E-value=1.01581e-06,
Score=126
Ontologies
GO
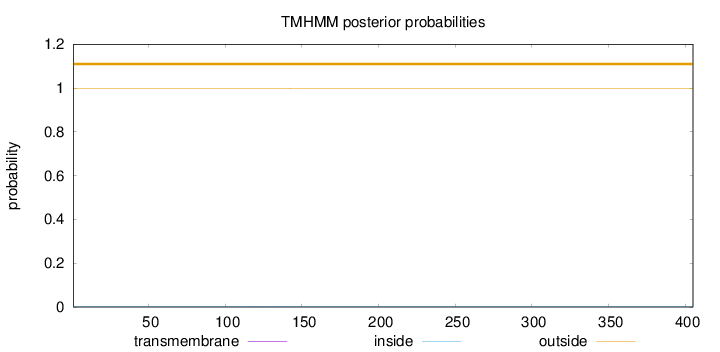
Topology
Length:
405
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00691
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00169
outside
1 - 405
Population Genetic Test Statistics
Pi
247.662686
Theta
166.029811
Tajima's D
1.694242
CLR
0.023153
CSRT
0.824558772061397
Interpretation
Uncertain
