Gene
KWMTBOMO14793
Pre Gene Modal
BGIBMGA012219
Annotation
PREDICTED:_pantothenate_kinase_3_isoform_X2_[Papilio_xuthus]
Location in the cell
Mitochondrial Reliability : 1.639
Sequence
CDS
ATGCGTCTTCATCGTTGTGAGGGCTGCCTAGCGGCGTGCGGCAGAGAACGCCGCGCCGCCCTCTCTTCAACCCCTAGACCCCTCATCAGGACTAAGCTATTGAAACGAGCGAGACGTAATCCGTACTCGCCTCCGCACGCAACGACAAAGGAACACTTCCCAGCCAAGATGGCGGAGTCCGCACCGCCCACGCCGCCCGACAATAACGCCCCACCGATGCCCTGGTTCGGCATGGACATTGGGGGCACGCTGACGAAACTGGTCTATTTCGAGCCCCGCGAGACGAACAGACGCGAACTCGACAAGGAGACCGAAGTCCTGAAGAATATCAGGAGATACTTGACGAGGAACTCGGCTTACGGGAAGACCGGGAGAAGGGACGTTCATCTGCAGATGGAGAACGTAACGATAAGAGGCCGCAGGGGCACGCTGCACTTCATAAGGTTCCCGACTTCGGAGGTGGGAGCTTTCCTCCAGCTGGCCCGCTCTAAGGGCATGGCGGATCTGGTGAACACAATCTACGCGACAGGGGGGGGCGCGTACAAGTTCGAGGAGGACTTCATTAAGGAGGTGAATATGCGTCTCTCAAAGATGGACGAACTAGACGCTCTGATCGCGGGCGTTTTGTACGTGGACTCTATGAATCCGAGGGAGTGCTACTATTGGGCGCCGCCAGACGAGCAGCCCGTGCACCAGCTGCCCAGCGACACGCCTCCTTACTTGGAAGCCGCCCTTAGTCAATACGTTCGCAAGCCCTACGACTTCTCCTGTCCCTACCCGTTCCTGTTGGTGAACATCGGCAGCGGAGTGTCCCTGCTTGTGGTCAACGGACCGGACAACTACTACAGAGTCAGCGGGACCAGTCTCGGCGGGGGGACGTTCCTGGGGCTGTGCTGTCTGCTGACCGGCTGCAGCAGCTTCGAGGAGGCGATATCTCTGGCGGCGTCCGGGGACAACACCACCGTGGACAAGCTGGTGCGCGACATATACGGCGGGGACTACGCGCGCTTCGGCTTGCCCGGGGACCTGGTGGCCAGCAGCTTCGGGCAGATGTGCTCCGCGGACCGAAGGGCCAAGGTGTCGAGGGAGGACCTGGCGCGGGCCACGCTGGTCACCATCACCAACAACATCGGCTCGATAGCGAGGCTGTGCGCTTGCAACGAGCGCGTCGAGAGGGTTGTATTTTGCGGTAACTTCCTACGAGTGAATCCGTTGTCGATGAAGCTTTTGTCGTACGCCATGTCCTACTGGTCGAGGGGAGCTTTGAAAGCACTGTTCTTGGAGCACGAGGGCTACTTTGGGGCCGTCGGCTGTCTCCTGCACCTCGACGCTGAGAACCACAAGAAGCTGCGGAACCGCACGCCACCGACGGGCGACGCCGCGCACGCAGACAGGTGA
Protein
MRLHRCEGCLAACGRERRAALSSTPRPLIRTKLLKRARRNPYSPPHATTKEHFPAKMAESAPPTPPDNNAPPMPWFGMDIGGTLTKLVYFEPRETNRRELDKETEVLKNIRRYLTRNSAYGKTGRRDVHLQMENVTIRGRRGTLHFIRFPTSEVGAFLQLARSKGMADLVNTIYATGGGAYKFEEDFIKEVNMRLSKMDELDALIAGVLYVDSMNPRECYYWAPPDEQPVHQLPSDTPPYLEAALSQYVRKPYDFSCPYPFLLVNIGSGVSLLVVNGPDNYYRVSGTSLGGGTFLGLCCLLTGCSSFEEAISLAASGDNTTVDKLVRDIYGGDYARFGLPGDLVASSFGQMCSADRRAKVSREDLARATLVTITNNIGSIARLCACNERVERVVFCGNFLRVNPLSMKLLSYAMSYWSRGALKALFLEHEGYFGAVGCLLHLDAENHKKLRNRTPPTGDAAHADR
Summary
Uniprot
A0A194Q0Y0
A0A2A4JD54
A0A194R4L7
A0A0C9PPJ8
A0A088AF52
A0A0C9R5A9
+ More
E2C1K4 A0A158NVH1 A0A0J7KZU3 A0A2A3ET07 E2AR36 A0A151ILP8 A0A1W4X754 A0A3L8D4Q6 A0A151JA08 A0A151I3G8 K7IZS2 A0A151WQ71 A0A067QMZ6 A0A1L8E019 A0A1Y1K1F9 A0A1W4WWB1 A0A1W4WWC1 A0A154PUU9 A0A1W4X5R2 A0A1L8DZI5 A0A1B0CMK8 A0A310SIU1 U5EQZ6 A0A026VW65 T1HR44 A0A2J7RPV8 E0VG17 A0A1L8DZH9 A0A2J7RPU5 A0A232EST4 A0A023EQU9 A0A0N7Z8Y2 Q16M65 A0A1Y1K6I0 A0A182FU31 A0A182U6K6 A0A2M4ABH6 A0A069DT88 Q7PVC2 T1E1F3 A0A336MV69 A0A182YPP9 A0A2M3Z3B8 A0A2M3Z3A6 A0A336MQ19 A0A2M4D0G2 A0A182NPN0 A0A2M4AAK1 A0A182WEE9 A0A084VGF8 W5JFZ1 A0A2M4BJ25 A0A182HH21 A0A182M1P4 A0A195FKU5 A0A0L7QYN5 A0A182IQY9 A0A182L4H4 A0A139WNF2 A0A182XKR4 A0A182PC53 A0A182VLS7 A0A182QVP7 A0A182R4H0 A0A0K8TNF6 A0A182K4R8 A0A0A9X5Z5 A0A139WNI0 V5GNM8 A0A1Q3FE95 A0A1Q3FE38 A0A1Q3FE72 A0A1Q3FDW0 A0A1Q3FE55 E9IWQ0 A0A1B6E196 A0A034VST6 A0A0K8WHC3 A0A034VTM6 A0A0K8V0B6 A0A1B6CCW4 A0A034VPE6 A0A0K8WE18 A0A0K8W8S2 U4USC3 A0A1B6EW86 A0A0Q5U5Q9 A0A0Q5U9C4 A0A0Q5UHH7 A0A1B6MIH1 A0A0R1DYZ3 Q8I0S6 A0A0R1DZ95 B4PF92
E2C1K4 A0A158NVH1 A0A0J7KZU3 A0A2A3ET07 E2AR36 A0A151ILP8 A0A1W4X754 A0A3L8D4Q6 A0A151JA08 A0A151I3G8 K7IZS2 A0A151WQ71 A0A067QMZ6 A0A1L8E019 A0A1Y1K1F9 A0A1W4WWB1 A0A1W4WWC1 A0A154PUU9 A0A1W4X5R2 A0A1L8DZI5 A0A1B0CMK8 A0A310SIU1 U5EQZ6 A0A026VW65 T1HR44 A0A2J7RPV8 E0VG17 A0A1L8DZH9 A0A2J7RPU5 A0A232EST4 A0A023EQU9 A0A0N7Z8Y2 Q16M65 A0A1Y1K6I0 A0A182FU31 A0A182U6K6 A0A2M4ABH6 A0A069DT88 Q7PVC2 T1E1F3 A0A336MV69 A0A182YPP9 A0A2M3Z3B8 A0A2M3Z3A6 A0A336MQ19 A0A2M4D0G2 A0A182NPN0 A0A2M4AAK1 A0A182WEE9 A0A084VGF8 W5JFZ1 A0A2M4BJ25 A0A182HH21 A0A182M1P4 A0A195FKU5 A0A0L7QYN5 A0A182IQY9 A0A182L4H4 A0A139WNF2 A0A182XKR4 A0A182PC53 A0A182VLS7 A0A182QVP7 A0A182R4H0 A0A0K8TNF6 A0A182K4R8 A0A0A9X5Z5 A0A139WNI0 V5GNM8 A0A1Q3FE95 A0A1Q3FE38 A0A1Q3FE72 A0A1Q3FDW0 A0A1Q3FE55 E9IWQ0 A0A1B6E196 A0A034VST6 A0A0K8WHC3 A0A034VTM6 A0A0K8V0B6 A0A1B6CCW4 A0A034VPE6 A0A0K8WE18 A0A0K8W8S2 U4USC3 A0A1B6EW86 A0A0Q5U5Q9 A0A0Q5U9C4 A0A0Q5UHH7 A0A1B6MIH1 A0A0R1DYZ3 Q8I0S6 A0A0R1DZ95 B4PF92
Pubmed
26354079
20798317
21347285
30249741
20075255
24845553
+ More
28004739 24508170 20566863 28648823 24945155 27129103 17510324 26334808 12364791 24330624 25244985 24438588 20920257 23761445 20966253 18362917 19820115 26369729 25401762 26823975 21282665 25348373 23537049 17994087 18057021 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
28004739 24508170 20566863 28648823 24945155 27129103 17510324 26334808 12364791 24330624 25244985 24438588 20920257 23761445 20966253 18362917 19820115 26369729 25401762 26823975 21282665 25348373 23537049 17994087 18057021 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
EMBL
KQ459580
KPI99242.1
NWSH01001892
PCG69779.1
KQ460685
KPJ12758.1
+ More
GBYB01003128 GBYB01012483 JAG72895.1 JAG82250.1 GBYB01003130 JAG72897.1 GL451937 EFN78180.1 ADTU01027159 LBMM01001695 KMQ95886.1 KZ288185 PBC34875.1 GL441869 EFN64106.1 KQ977104 KYN05812.1 QOIP01000013 RLU15370.1 KQ979348 KYN21857.1 KQ976494 KYM83289.1 KQ982850 KYQ49953.1 KK853149 KDR10674.1 GFDF01002192 JAV11892.1 GEZM01095875 GEZM01095870 JAV55284.1 KQ435180 KZC14980.1 GFDF01002247 JAV11837.1 AJWK01018841 AJWK01018842 AJWK01018843 KQ765222 OAD54064.1 GANO01004011 JAB55860.1 KK107796 EZA47746.1 ACPB03022348 ACPB03022349 NEVH01001355 PNF42863.1 DS235131 EEB12323.1 GFDF01002248 JAV11836.1 PNF42862.1 NNAY01002392 OXU21377.1 GAPW01002103 JAC11495.1 GDKW01002075 JAI54520.1 CH477878 EAT35421.1 GEZM01095874 GEZM01095873 JAV55285.1 GGFK01004823 MBW38144.1 GBGD01001724 JAC87165.1 AAAB01008986 EAA00064.4 GALA01001729 JAA93123.1 UFQT01002092 SSX32643.1 GGFM01002242 MBW22993.1 GGFM01002258 MBW23009.1 SSX32644.1 GGFL01006410 MBW70588.1 GGFK01004441 MBW37762.1 ATLV01012877 KE524821 KFB37052.1 ADMH02001328 ETN62981.1 GGFJ01003842 MBW52983.1 APCN01002375 AXCM01002269 KQ981512 KYN40882.1 KQ414688 KOC63672.1 KQ971311 KYB29462.1 AXCN02001648 GDAI01001704 JAI15899.1 GBHO01029372 GBHO01029371 GBRD01008230 GBRD01008225 GBRD01008224 GBRD01008222 GDHC01021899 GDHC01015034 JAG14232.1 JAG14233.1 JAG57591.1 JAP96729.1 JAQ03595.1 KYB29463.1 GALX01002733 GALX01002732 JAB65734.1 GFDL01009169 JAV25876.1 GFDL01009220 JAV25825.1 GFDL01009206 JAV25839.1 GFDL01009291 JAV25754.1 GFDL01009205 JAV25840.1 GL766597 EFZ14992.1 GEDC01018064 GEDC01005631 JAS19234.1 JAS31667.1 GAKP01013790 GAKP01013788 JAC45162.1 GDHF01009362 GDHF01001850 JAI42952.1 JAI50464.1 GAKP01013789 GAKP01013787 JAC45165.1 GDHF01020023 GDHF01010215 JAI32291.1 JAI42099.1 GEDC01029352 GEDC01026026 JAS07946.1 JAS11272.1 GAKP01013786 GAKP01013785 JAC45166.1 GDHF01003209 JAI49105.1 GDHF01004768 JAI47546.1 KB632345 ERL93076.1 GECZ01027557 GECZ01023082 GECZ01015814 GECZ01005046 JAS42212.1 JAS46687.1 JAS53955.1 JAS64723.1 CH954178 KQS44309.1 KQS44308.1 KQS44310.1 KQS44307.1 GEBQ01004264 JAT35713.1 CM000159 KRK02278.1 AE014296 BT001335 AAN12127.1 AAN12128.1 AAN71090.1 KRK02277.1 EDW95178.1
GBYB01003128 GBYB01012483 JAG72895.1 JAG82250.1 GBYB01003130 JAG72897.1 GL451937 EFN78180.1 ADTU01027159 LBMM01001695 KMQ95886.1 KZ288185 PBC34875.1 GL441869 EFN64106.1 KQ977104 KYN05812.1 QOIP01000013 RLU15370.1 KQ979348 KYN21857.1 KQ976494 KYM83289.1 KQ982850 KYQ49953.1 KK853149 KDR10674.1 GFDF01002192 JAV11892.1 GEZM01095875 GEZM01095870 JAV55284.1 KQ435180 KZC14980.1 GFDF01002247 JAV11837.1 AJWK01018841 AJWK01018842 AJWK01018843 KQ765222 OAD54064.1 GANO01004011 JAB55860.1 KK107796 EZA47746.1 ACPB03022348 ACPB03022349 NEVH01001355 PNF42863.1 DS235131 EEB12323.1 GFDF01002248 JAV11836.1 PNF42862.1 NNAY01002392 OXU21377.1 GAPW01002103 JAC11495.1 GDKW01002075 JAI54520.1 CH477878 EAT35421.1 GEZM01095874 GEZM01095873 JAV55285.1 GGFK01004823 MBW38144.1 GBGD01001724 JAC87165.1 AAAB01008986 EAA00064.4 GALA01001729 JAA93123.1 UFQT01002092 SSX32643.1 GGFM01002242 MBW22993.1 GGFM01002258 MBW23009.1 SSX32644.1 GGFL01006410 MBW70588.1 GGFK01004441 MBW37762.1 ATLV01012877 KE524821 KFB37052.1 ADMH02001328 ETN62981.1 GGFJ01003842 MBW52983.1 APCN01002375 AXCM01002269 KQ981512 KYN40882.1 KQ414688 KOC63672.1 KQ971311 KYB29462.1 AXCN02001648 GDAI01001704 JAI15899.1 GBHO01029372 GBHO01029371 GBRD01008230 GBRD01008225 GBRD01008224 GBRD01008222 GDHC01021899 GDHC01015034 JAG14232.1 JAG14233.1 JAG57591.1 JAP96729.1 JAQ03595.1 KYB29463.1 GALX01002733 GALX01002732 JAB65734.1 GFDL01009169 JAV25876.1 GFDL01009220 JAV25825.1 GFDL01009206 JAV25839.1 GFDL01009291 JAV25754.1 GFDL01009205 JAV25840.1 GL766597 EFZ14992.1 GEDC01018064 GEDC01005631 JAS19234.1 JAS31667.1 GAKP01013790 GAKP01013788 JAC45162.1 GDHF01009362 GDHF01001850 JAI42952.1 JAI50464.1 GAKP01013789 GAKP01013787 JAC45165.1 GDHF01020023 GDHF01010215 JAI32291.1 JAI42099.1 GEDC01029352 GEDC01026026 JAS07946.1 JAS11272.1 GAKP01013786 GAKP01013785 JAC45166.1 GDHF01003209 JAI49105.1 GDHF01004768 JAI47546.1 KB632345 ERL93076.1 GECZ01027557 GECZ01023082 GECZ01015814 GECZ01005046 JAS42212.1 JAS46687.1 JAS53955.1 JAS64723.1 CH954178 KQS44309.1 KQS44308.1 KQS44310.1 KQS44307.1 GEBQ01004264 JAT35713.1 CM000159 KRK02278.1 AE014296 BT001335 AAN12127.1 AAN12128.1 AAN71090.1 KRK02277.1 EDW95178.1
Proteomes
UP000053268
UP000218220
UP000053240
UP000005203
UP000008237
UP000005205
+ More
UP000036403 UP000242457 UP000000311 UP000078542 UP000192223 UP000279307 UP000078492 UP000078540 UP000002358 UP000075809 UP000027135 UP000076502 UP000092461 UP000053097 UP000015103 UP000235965 UP000009046 UP000215335 UP000008820 UP000069272 UP000075902 UP000007062 UP000076408 UP000075884 UP000075920 UP000030765 UP000000673 UP000075840 UP000075883 UP000078541 UP000053825 UP000075880 UP000075882 UP000007266 UP000076407 UP000075885 UP000075903 UP000075886 UP000075900 UP000075881 UP000030742 UP000008711 UP000002282 UP000000803
UP000036403 UP000242457 UP000000311 UP000078542 UP000192223 UP000279307 UP000078492 UP000078540 UP000002358 UP000075809 UP000027135 UP000076502 UP000092461 UP000053097 UP000015103 UP000235965 UP000009046 UP000215335 UP000008820 UP000069272 UP000075902 UP000007062 UP000076408 UP000075884 UP000075920 UP000030765 UP000000673 UP000075840 UP000075883 UP000078541 UP000053825 UP000075880 UP000075882 UP000007266 UP000076407 UP000075885 UP000075903 UP000075886 UP000075900 UP000075881 UP000030742 UP000008711 UP000002282 UP000000803
PRIDE
ProteinModelPortal
A0A194Q0Y0
A0A2A4JD54
A0A194R4L7
A0A0C9PPJ8
A0A088AF52
A0A0C9R5A9
+ More
E2C1K4 A0A158NVH1 A0A0J7KZU3 A0A2A3ET07 E2AR36 A0A151ILP8 A0A1W4X754 A0A3L8D4Q6 A0A151JA08 A0A151I3G8 K7IZS2 A0A151WQ71 A0A067QMZ6 A0A1L8E019 A0A1Y1K1F9 A0A1W4WWB1 A0A1W4WWC1 A0A154PUU9 A0A1W4X5R2 A0A1L8DZI5 A0A1B0CMK8 A0A310SIU1 U5EQZ6 A0A026VW65 T1HR44 A0A2J7RPV8 E0VG17 A0A1L8DZH9 A0A2J7RPU5 A0A232EST4 A0A023EQU9 A0A0N7Z8Y2 Q16M65 A0A1Y1K6I0 A0A182FU31 A0A182U6K6 A0A2M4ABH6 A0A069DT88 Q7PVC2 T1E1F3 A0A336MV69 A0A182YPP9 A0A2M3Z3B8 A0A2M3Z3A6 A0A336MQ19 A0A2M4D0G2 A0A182NPN0 A0A2M4AAK1 A0A182WEE9 A0A084VGF8 W5JFZ1 A0A2M4BJ25 A0A182HH21 A0A182M1P4 A0A195FKU5 A0A0L7QYN5 A0A182IQY9 A0A182L4H4 A0A139WNF2 A0A182XKR4 A0A182PC53 A0A182VLS7 A0A182QVP7 A0A182R4H0 A0A0K8TNF6 A0A182K4R8 A0A0A9X5Z5 A0A139WNI0 V5GNM8 A0A1Q3FE95 A0A1Q3FE38 A0A1Q3FE72 A0A1Q3FDW0 A0A1Q3FE55 E9IWQ0 A0A1B6E196 A0A034VST6 A0A0K8WHC3 A0A034VTM6 A0A0K8V0B6 A0A1B6CCW4 A0A034VPE6 A0A0K8WE18 A0A0K8W8S2 U4USC3 A0A1B6EW86 A0A0Q5U5Q9 A0A0Q5U9C4 A0A0Q5UHH7 A0A1B6MIH1 A0A0R1DYZ3 Q8I0S6 A0A0R1DZ95 B4PF92
E2C1K4 A0A158NVH1 A0A0J7KZU3 A0A2A3ET07 E2AR36 A0A151ILP8 A0A1W4X754 A0A3L8D4Q6 A0A151JA08 A0A151I3G8 K7IZS2 A0A151WQ71 A0A067QMZ6 A0A1L8E019 A0A1Y1K1F9 A0A1W4WWB1 A0A1W4WWC1 A0A154PUU9 A0A1W4X5R2 A0A1L8DZI5 A0A1B0CMK8 A0A310SIU1 U5EQZ6 A0A026VW65 T1HR44 A0A2J7RPV8 E0VG17 A0A1L8DZH9 A0A2J7RPU5 A0A232EST4 A0A023EQU9 A0A0N7Z8Y2 Q16M65 A0A1Y1K6I0 A0A182FU31 A0A182U6K6 A0A2M4ABH6 A0A069DT88 Q7PVC2 T1E1F3 A0A336MV69 A0A182YPP9 A0A2M3Z3B8 A0A2M3Z3A6 A0A336MQ19 A0A2M4D0G2 A0A182NPN0 A0A2M4AAK1 A0A182WEE9 A0A084VGF8 W5JFZ1 A0A2M4BJ25 A0A182HH21 A0A182M1P4 A0A195FKU5 A0A0L7QYN5 A0A182IQY9 A0A182L4H4 A0A139WNF2 A0A182XKR4 A0A182PC53 A0A182VLS7 A0A182QVP7 A0A182R4H0 A0A0K8TNF6 A0A182K4R8 A0A0A9X5Z5 A0A139WNI0 V5GNM8 A0A1Q3FE95 A0A1Q3FE38 A0A1Q3FE72 A0A1Q3FDW0 A0A1Q3FE55 E9IWQ0 A0A1B6E196 A0A034VST6 A0A0K8WHC3 A0A034VTM6 A0A0K8V0B6 A0A1B6CCW4 A0A034VPE6 A0A0K8WE18 A0A0K8W8S2 U4USC3 A0A1B6EW86 A0A0Q5U5Q9 A0A0Q5U9C4 A0A0Q5UHH7 A0A1B6MIH1 A0A0R1DYZ3 Q8I0S6 A0A0R1DZ95 B4PF92
PDB
2I7N
E-value=9.4475e-115,
Score=1058
Ontologies
PATHWAY
GO
GO:0005524
GO:0015937
GO:0004594
GO:0031514
GO:0048870
GO:0032465
GO:0007476
GO:0006979
GO:0048477
GO:0045475
GO:0007283
GO:0005886
GO:0071480
GO:0040011
GO:0000278
GO:0051321
GO:0061024
GO:0070328
GO:0005737
GO:0005819
GO:0007059
GO:0008270
GO:0016701
GO:0006355
GO:0003824
GO:0008152
GO:0016491
GO:0015930
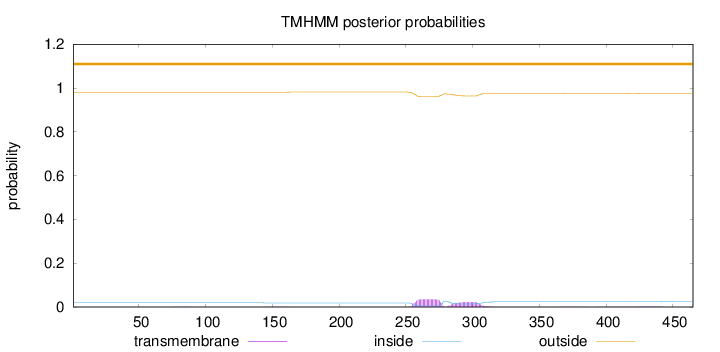
Topology
Length:
465
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.26262
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01891
outside
1 - 465
Population Genetic Test Statistics
Pi
247.320109
Theta
165.083636
Tajima's D
1.588725
CLR
0.23748
CSRT
0.808109594520274
Interpretation
Uncertain
