Pre Gene Modal
BGIBMGA012220
Annotation
PREDICTED:_tetratricopeptide_repeat_protein_1-like_[Amyelois_transitella]
Full name
Tetratricopeptide repeat protein 1
Location in the cell
Nuclear Reliability : 3.413
Sequence
CDS
ATGAATTTGTTTTGTGCGAATACATGGCTTCAAAATACGTGCAAAACTTCCAATGAGGCTGTAATTAATGACATCACAAGGGATTTGGAAACATCGCTCAATACTAACGAAAATACTGAAGGATATGTCATTCAGCCTGGTACTAGTGAGGAACAATGCTTTTCTACAGAAACAGCGAAAAATCATACAGAACTACCCAAAGATGCAGAATTCGACGCTGACCATAACGCATCAGACAACGAAGATGACAAAGAAAGCGATACAGATTCTATAGATGAAGAACATCTAAAAGATGACGAGGTAGCACTAACGGATGACCAGAAAGCAGAAAGGCTGATCATGGCCGAGGAATTGAAACAGGCCGGAAACAATTCTTACAAATGTGGAGAGTATGAGAGAAGTGTAGAGAAATACACCGAAGGTTTGAGATTATGTCCTCTTCAATTCGACAAAGTACGTGCGATTCTATATTGCAACAGGAGTGCGGCTAGGATGAAATTGAAAAAGTATCAAAAGTCAATCAAGGACTGCACTAAAGCACTGGAACTCGATGACAAATATGTCAAAGCGTACTTAAGGCGTGCCTCGTCCAACGAGCTATTGGAAAAGTATCAAGACGCCTTGAGCGATTATGAGAAGGCTCTAGAATTGGACCCGTCTTTGGAGGAGACCAGGAAAGCCGTGGTCCGCCTGAAGCCGATCGTGGAAGCCAAGAATGAGGAAATGAAGAAAGAGATGCTGGGGAAACTGAAGGATCTGGGGAACATGATACTGAAGCCGTTCGGTCTGTCCACGGAGAACTTTAATATGGAACAGGACCCGGAGACGAAAGGATACAAAATTAATTTTAAGCAATAA
Protein
MNLFCANTWLQNTCKTSNEAVINDITRDLETSLNTNENTEGYVIQPGTSEEQCFSTETAKNHTELPKDAEFDADHNASDNEDDKESDTDSIDEEHLKDDEVALTDDQKAERLIMAEELKQAGNNSYKCGEYERSVEKYTEGLRLCPLQFDKVRAILYCNRSAARMKLKKYQKSIKDCTKALELDDKYVKAYLRRASSNELLEKYQDALSDYEKALELDPSLEETRKAVVRLKPIVEAKNEEMKKEMLGKLKDLGNMILKPFGLSTENFNMEQDPETKGYKINFKQ
Summary
Subunit
Interacts with the GAP domain of NF1. Interacts (via TPR repeats) with HSP90AA1 and HSPA8.
Keywords
Complete proteome
Phosphoprotein
Reference proteome
Repeat
TPR repeat
Feature
chain Tetratricopeptide repeat protein 1
Uniprot
H9JRR0
A0A2A4JWR3
A0A212EZE9
A0A194Q769
A0A182NFU3
A0A0K8TI18
+ More
A0A0A9X6M6 A0A151X902 A0A182QP00 A0A151J3K4 E9J750 A0A026WE21 U5EYP7 A0A182HJK0 A0A182XKE4 Q7Q9B6 A0A151IAH6 A0A182J2Z7 A0A182KWQ5 A0A195B5R0 A0A158P136 A0A2M4ARH5 F4W999 A0A0C9RRQ3 A0A2J7RFQ7 A0A182TKP5 B0XDM8 A0A154PN89 A0A1W4W4R3 A0A182UZQ2 A0A182XXA8 E2B4B8 A0A1Q3F4A1 A0A310SKA6 A0A067RS22 A0A0L7RHV7 D6X2V6 E2AM74 A0A3L8DQ18 A0A232EVR0 A0A1S3JTY5 U4UPL4 N6U2U0 A0A0M8ZSB9 A0A1B6CG96 E0VUW9 A0A1I8M6X4 T1PBC3 A0A3S2NR27 A0A1Q3F440 A0A1Y1MSK3 B4NL99 A0A2A3EI17 A0A1B6MM46 Q16V27 A0A0L0BN82 A0A1B6JMC0 A0A2G8L654 A0A1J1IRN5 A0A0P4VMF2 R4FM29 A0A088ACQ9 A0A2Y9DC46 L8IG12 A0A1Z5L7K9 A0A2Y9ELB8 A0A341BUJ2 U3K2M9 Q3ZBR5 A0A2U3UZ02 A0A2Y9MX63 A0A1B6EQ97 A0A0V0G5W1 A0A287AVJ6 R4WPA2 W5PUM2
A0A0A9X6M6 A0A151X902 A0A182QP00 A0A151J3K4 E9J750 A0A026WE21 U5EYP7 A0A182HJK0 A0A182XKE4 Q7Q9B6 A0A151IAH6 A0A182J2Z7 A0A182KWQ5 A0A195B5R0 A0A158P136 A0A2M4ARH5 F4W999 A0A0C9RRQ3 A0A2J7RFQ7 A0A182TKP5 B0XDM8 A0A154PN89 A0A1W4W4R3 A0A182UZQ2 A0A182XXA8 E2B4B8 A0A1Q3F4A1 A0A310SKA6 A0A067RS22 A0A0L7RHV7 D6X2V6 E2AM74 A0A3L8DQ18 A0A232EVR0 A0A1S3JTY5 U4UPL4 N6U2U0 A0A0M8ZSB9 A0A1B6CG96 E0VUW9 A0A1I8M6X4 T1PBC3 A0A3S2NR27 A0A1Q3F440 A0A1Y1MSK3 B4NL99 A0A2A3EI17 A0A1B6MM46 Q16V27 A0A0L0BN82 A0A1B6JMC0 A0A2G8L654 A0A1J1IRN5 A0A0P4VMF2 R4FM29 A0A088ACQ9 A0A2Y9DC46 L8IG12 A0A1Z5L7K9 A0A2Y9ELB8 A0A341BUJ2 U3K2M9 Q3ZBR5 A0A2U3UZ02 A0A2Y9MX63 A0A1B6EQ97 A0A0V0G5W1 A0A287AVJ6 R4WPA2 W5PUM2
Pubmed
19121390
22118469
26354079
25401762
21282665
24508170
+ More
12364791 14747013 17210077 20966253 21347285 21719571 25244985 20798317 24845553 18362917 19820115 30249741 28648823 23537049 20566863 25315136 28004739 17994087 17510324 26108605 29023486 27129103 22751099 28528879 30723633 23691247 20809919
12364791 14747013 17210077 20966253 21347285 21719571 25244985 20798317 24845553 18362917 19820115 30249741 28648823 23537049 20566863 25315136 28004739 17994087 17510324 26108605 29023486 27129103 22751099 28528879 30723633 23691247 20809919
EMBL
BABH01040834
NWSH01000437
PCG76441.1
AGBW02011291
OWR46879.1
KQ459580
+ More
KPI99240.1 GBRD01000583 JAG65238.1 GBHO01027217 JAG16387.1 KQ982402 KYQ56842.1 AXCN02000248 KQ980275 KYN17026.1 GL768421 EFZ11392.1 KK107293 EZA53279.1 GANO01001888 JAB57983.1 APCN01002178 AAAB01008904 EAA09638.5 KQ978223 KYM96268.1 KQ976598 KYM79597.1 ADTU01001043 GGFK01010048 MBW43369.1 GL888002 EGI69281.1 GBYB01011230 JAG80997.1 NEVH01004410 PNF39669.1 DS232769 EDS45536.1 KQ434960 KZC12690.1 GL445549 EFN89466.1 GFDL01012692 JAV22353.1 KQ760478 OAD60236.1 KK852458 KDR23545.1 KQ414590 KOC70399.1 KQ971372 EFA10651.2 GL440715 EFN65480.1 QOIP01000005 RLU22497.1 NNAY01001968 OXU22425.1 KB632309 ERL92096.1 APGK01044467 APGK01044468 KB741026 ENN74931.1 KQ435876 KOX70135.1 GEDC01024836 JAS12462.1 DS235797 EEB17175.1 KA646061 AFP60690.1 RSAL01000132 RVE46371.1 GFDL01012721 JAV22324.1 GEZM01025399 JAV87475.1 CH964272 EDW84302.1 KZ288234 PBC31433.1 GEBQ01002967 JAT37010.1 CH477605 EAT38403.1 JRES01001621 KNC21477.1 GECU01007381 JAT00326.1 MRZV01000202 PIK55741.1 CVRI01000057 CRL02386.1 GDKW01000273 JAI56322.1 ACPB03018838 GAHY01001153 JAA76357.1 JH881299 ELR55138.1 GFJQ02003563 JAW03407.1 AGTO01020935 BC103153 GECZ01029654 JAS40115.1 GECL01002686 JAP03438.1 AEMK02000102 DQIR01257876 HDC13354.1 AK417491 BAN20706.1 AMGL01093842
KPI99240.1 GBRD01000583 JAG65238.1 GBHO01027217 JAG16387.1 KQ982402 KYQ56842.1 AXCN02000248 KQ980275 KYN17026.1 GL768421 EFZ11392.1 KK107293 EZA53279.1 GANO01001888 JAB57983.1 APCN01002178 AAAB01008904 EAA09638.5 KQ978223 KYM96268.1 KQ976598 KYM79597.1 ADTU01001043 GGFK01010048 MBW43369.1 GL888002 EGI69281.1 GBYB01011230 JAG80997.1 NEVH01004410 PNF39669.1 DS232769 EDS45536.1 KQ434960 KZC12690.1 GL445549 EFN89466.1 GFDL01012692 JAV22353.1 KQ760478 OAD60236.1 KK852458 KDR23545.1 KQ414590 KOC70399.1 KQ971372 EFA10651.2 GL440715 EFN65480.1 QOIP01000005 RLU22497.1 NNAY01001968 OXU22425.1 KB632309 ERL92096.1 APGK01044467 APGK01044468 KB741026 ENN74931.1 KQ435876 KOX70135.1 GEDC01024836 JAS12462.1 DS235797 EEB17175.1 KA646061 AFP60690.1 RSAL01000132 RVE46371.1 GFDL01012721 JAV22324.1 GEZM01025399 JAV87475.1 CH964272 EDW84302.1 KZ288234 PBC31433.1 GEBQ01002967 JAT37010.1 CH477605 EAT38403.1 JRES01001621 KNC21477.1 GECU01007381 JAT00326.1 MRZV01000202 PIK55741.1 CVRI01000057 CRL02386.1 GDKW01000273 JAI56322.1 ACPB03018838 GAHY01001153 JAA76357.1 JH881299 ELR55138.1 GFJQ02003563 JAW03407.1 AGTO01020935 BC103153 GECZ01029654 JAS40115.1 GECL01002686 JAP03438.1 AEMK02000102 DQIR01257876 HDC13354.1 AK417491 BAN20706.1 AMGL01093842
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000075884
UP000075809
+ More
UP000075886 UP000078492 UP000053097 UP000075840 UP000076407 UP000007062 UP000078542 UP000075880 UP000075882 UP000078540 UP000005205 UP000007755 UP000235965 UP000075902 UP000002320 UP000076502 UP000192223 UP000075903 UP000076408 UP000008237 UP000027135 UP000053825 UP000007266 UP000000311 UP000279307 UP000215335 UP000085678 UP000030742 UP000019118 UP000053105 UP000009046 UP000095301 UP000283053 UP000007798 UP000242457 UP000008820 UP000037069 UP000230750 UP000183832 UP000015103 UP000005203 UP000248480 UP000248484 UP000252040 UP000016665 UP000009136 UP000245320 UP000248483 UP000008227 UP000002356
UP000075886 UP000078492 UP000053097 UP000075840 UP000076407 UP000007062 UP000078542 UP000075880 UP000075882 UP000078540 UP000005205 UP000007755 UP000235965 UP000075902 UP000002320 UP000076502 UP000192223 UP000075903 UP000076408 UP000008237 UP000027135 UP000053825 UP000007266 UP000000311 UP000279307 UP000215335 UP000085678 UP000030742 UP000019118 UP000053105 UP000009046 UP000095301 UP000283053 UP000007798 UP000242457 UP000008820 UP000037069 UP000230750 UP000183832 UP000015103 UP000005203 UP000248480 UP000248484 UP000252040 UP000016665 UP000009136 UP000245320 UP000248483 UP000008227 UP000002356
Interpro
Gene 3D
ProteinModelPortal
H9JRR0
A0A2A4JWR3
A0A212EZE9
A0A194Q769
A0A182NFU3
A0A0K8TI18
+ More
A0A0A9X6M6 A0A151X902 A0A182QP00 A0A151J3K4 E9J750 A0A026WE21 U5EYP7 A0A182HJK0 A0A182XKE4 Q7Q9B6 A0A151IAH6 A0A182J2Z7 A0A182KWQ5 A0A195B5R0 A0A158P136 A0A2M4ARH5 F4W999 A0A0C9RRQ3 A0A2J7RFQ7 A0A182TKP5 B0XDM8 A0A154PN89 A0A1W4W4R3 A0A182UZQ2 A0A182XXA8 E2B4B8 A0A1Q3F4A1 A0A310SKA6 A0A067RS22 A0A0L7RHV7 D6X2V6 E2AM74 A0A3L8DQ18 A0A232EVR0 A0A1S3JTY5 U4UPL4 N6U2U0 A0A0M8ZSB9 A0A1B6CG96 E0VUW9 A0A1I8M6X4 T1PBC3 A0A3S2NR27 A0A1Q3F440 A0A1Y1MSK3 B4NL99 A0A2A3EI17 A0A1B6MM46 Q16V27 A0A0L0BN82 A0A1B6JMC0 A0A2G8L654 A0A1J1IRN5 A0A0P4VMF2 R4FM29 A0A088ACQ9 A0A2Y9DC46 L8IG12 A0A1Z5L7K9 A0A2Y9ELB8 A0A341BUJ2 U3K2M9 Q3ZBR5 A0A2U3UZ02 A0A2Y9MX63 A0A1B6EQ97 A0A0V0G5W1 A0A287AVJ6 R4WPA2 W5PUM2
A0A0A9X6M6 A0A151X902 A0A182QP00 A0A151J3K4 E9J750 A0A026WE21 U5EYP7 A0A182HJK0 A0A182XKE4 Q7Q9B6 A0A151IAH6 A0A182J2Z7 A0A182KWQ5 A0A195B5R0 A0A158P136 A0A2M4ARH5 F4W999 A0A0C9RRQ3 A0A2J7RFQ7 A0A182TKP5 B0XDM8 A0A154PN89 A0A1W4W4R3 A0A182UZQ2 A0A182XXA8 E2B4B8 A0A1Q3F4A1 A0A310SKA6 A0A067RS22 A0A0L7RHV7 D6X2V6 E2AM74 A0A3L8DQ18 A0A232EVR0 A0A1S3JTY5 U4UPL4 N6U2U0 A0A0M8ZSB9 A0A1B6CG96 E0VUW9 A0A1I8M6X4 T1PBC3 A0A3S2NR27 A0A1Q3F440 A0A1Y1MSK3 B4NL99 A0A2A3EI17 A0A1B6MM46 Q16V27 A0A0L0BN82 A0A1B6JMC0 A0A2G8L654 A0A1J1IRN5 A0A0P4VMF2 R4FM29 A0A088ACQ9 A0A2Y9DC46 L8IG12 A0A1Z5L7K9 A0A2Y9ELB8 A0A341BUJ2 U3K2M9 Q3ZBR5 A0A2U3UZ02 A0A2Y9MX63 A0A1B6EQ97 A0A0V0G5W1 A0A287AVJ6 R4WPA2 W5PUM2
PDB
4JA9
E-value=4.444e-16,
Score=205
Ontologies
GO
PANTHER
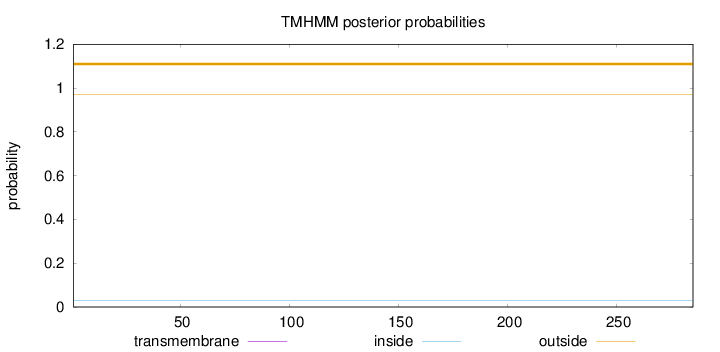
Topology
Length:
285
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00018
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03111
outside
1 - 285
Population Genetic Test Statistics
Pi
296.599004
Theta
160.019729
Tajima's D
3.103547
CLR
0.172289
CSRT
0.983300834958252
Interpretation
Uncertain
