Gene
KWMTBOMO14790 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012221
Annotation
transferin_[Helicoverpa_armigera]
Full name
Transferrin
Location in the cell
Extracellular Reliability : 1.805 PlasmaMembrane Reliability : 1.924
Sequence
CDS
ATGTTCGCGAAGCTTCTCATTCTATGCCTCGTGTTCGGCGCTAGCCGTGCCCAAAGTCATCGTGTGTGCATTTTGTCATCTTCATCGTCACTATGCCAGAGTTTAGACAAAGACGGGAGCCAGGCGACGTGCAGCAGAGTGGAAACTAGAATTGACTGCGCTTTGAGACTGGCCCGCGGAGAAGCGGACATTGGCTTCTTCACCGAGGAGGAGCTACTGTTATTATCACAGCAACAGCCAAACGACAATAGGGTCATCGCAACCGTCAGAGACGTCAGTAGATTGGAGACGTTCTCCTTCGAAGCTGTGGCTGTAGTTCCGAATAACCACACCGGAGGACTTGATGGGTTGCGTGGTGGTAGATATTGTCATCCCGGATTCGACCAGTCCGACCTGAGATGGTCCCCAAGAGTGCTCAAGACTTTGGAAGAAGTGGCCGCGCGCACCGACCGTTGCCCCGACGTGGACACGAACAGAAAAACGGCCGAGGAAATCGAAGTGGAAACATTAAGCTCCTTCTTCTCAGCTGCCTGCAGACCTGGGACCTGGAGTTCAAACTCTACAGTTGACGCCGACTTAAAAACGCGTTTCCCCTCTCTGTGCTCCCTATGCGGTGGGAGCTCCGGCTGCACAGGATACAGCATAGACATGGGCATCAGCGTCGCCGGAGTCAATAACCAGAACCGCCACATCCAGGCGTTGGAGTGCTTGAGGGTCAGCACGAATGACAGCGTCCCCACAGTGGCGTACGCGGCCTGGCTTCACGTCCGGAACTATTTTAATATCCGCAATCCACAAGACGCAGCCTCATTCTCCCTGCTGTGTCCGGATGGAACTCTTCAGGCGTTGACCCCTGAGATCTTGAGTAACCCTGTGGCACCTTGTTCGTTCGTGAAACAACCCTGGGGAGCTATTGTCGCGTCTACGGCTGCAGCTTCCACTGTCCTCACCAATCTCCAGGCCTGGTGGCCGACTGGAACTGATCCCGGAGCGAACTCTTGGCAGTCGGTATTGTTCAGCGGTCTCATTGGGGGCGCGAACGCGAGGGTTGTCTTCGAGCAAGCCCCCATTGCTCCAGCCAATTATACTGCTCCGATACGCAGCATTCCCGCTATAGACTCAACGGCGTCCTGTCTTCCGGCTCAACGCTGGTGTACCACGAACTTAAACGAGCAAACCAAGTGCTCTTGGGTTAGAGCATCAGCCTACACCCTCGGAATTCAGCCGGCAATCTCCTGTCAGCAGAGGAGCTCGGTCTTCGACTGCCTCACCGACATCAAGGAATCAAGAGCCGATTTTATTGCCACTCCCTCCAATTATGGATATATCGCAAGGCAACATTACCGCCTCTCGCCCGTGAAACTGGTTCAGAACACCAGATCTGGAGCGTCTCGGGTCGCGGCCTTCGTCAAGGAAACGGCCGCAGCGCAGGAGAACATCACGCGCTTCGAGAACCTTCGAGGAAAGAAGGCCTGCTTCCCAGAATTTGGAGGGCTTGCGTACGTCGCGTTCGTGCGCACCGCCCAGCTGCGGGGCGTGATCAGCGAGACAGAATGCGATTATGTGAAAGCTGTGGGAGAGTTTTTCGACGGCGCCTGCGCCCCGGGAGCATTGGACGCCTCTCACACCATCACGGAAACCACCTTCAACGCGACCTCGTTGTGTTCTGTATGCAGGCCACTCATCCCCACACCTGATAACTTTACCTGCGCTTATGATTATACGAACGTTTATTATGGCAACAACGGTACTCTAGCGTGTCTGAACGATCCGACCACGGACATCGCAGTGCTGAGCACGGAGAACATCAATGTTCACTTACAAGCGTTAGGACTGCAAGCCAATCAGTTCCGTGCCCTCTGTCAGAACAACACGCTCGCCGCGACGCCTGGCGTGTTCATTGACGACAACTGTCTCCTAGCTTACGTTGTCGACGCGGAGGTTCTGTCCAGAAGAGGCGACCCGCTGAACAACGCTTTGAACGTTTTATTCGATTCGCTGGACGCGTACTTCGGTTACAACACGGCGGCCGCGAACCAGCTCATCAATTTGGATTTGTTCTCGTCTTTCAACGGGATCAGCGATTTGTTGTTCAAGGACTCGACTATAGGCCTAACGGAGCCCACGGGCGAGGCTCCTCACGAACCCGCCAGGAACTACATGGAATTGTTCAGACACTTGGACGCGTGCAGCCGGGCGACCCCACAAGTGCCCGATATAGCGAACCGCGCCGCCTTCTCCGTTATAACGATGCTCGTTATGAGTTTCTTAACGAGATTTGTCGTTTATTAA
Protein
MFAKLLILCLVFGASRAQSHRVCILSSSSSLCQSLDKDGSQATCSRVETRIDCALRLARGEADIGFFTEEELLLLSQQQPNDNRVIATVRDVSRLETFSFEAVAVVPNNHTGGLDGLRGGRYCHPGFDQSDLRWSPRVLKTLEEVAARTDRCPDVDTNRKTAEEIEVETLSSFFSAACRPGTWSSNSTVDADLKTRFPSLCSLCGGSSGCTGYSIDMGISVAGVNNQNRHIQALECLRVSTNDSVPTVAYAAWLHVRNYFNIRNPQDAASFSLLCPDGTLQALTPEILSNPVAPCSFVKQPWGAIVASTAAASTVLTNLQAWWPTGTDPGANSWQSVLFSGLIGGANARVVFEQAPIAPANYTAPIRSIPAIDSTASCLPAQRWCTTNLNEQTKCSWVRASAYTLGIQPAISCQQRSSVFDCLTDIKESRADFIATPSNYGYIARQHYRLSPVKLVQNTRSGASRVAAFVKETAAAQENITRFENLRGKKACFPEFGGLAYVAFVRTAQLRGVISETECDYVKAVGEFFDGACAPGALDASHTITETTFNATSLCSVCRPLIPTPDNFTCAYDYTNVYYGNNGTLACLNDPTTDIAVLSTENINVHLQALGLQANQFRALCQNNTLAATPGVFIDDNCLLAYVVDAEVLSRRGDPLNNALNVLFDSLDAYFGYNTAAANQLINLDLFSSFNGISDLLFKDSTIGLTEPTGEAPHEPARNYMELFRHLDACSRATPQVPDIANRAAFSVITMLVMSFLTRFVVY
Summary
Description
Transferrins are iron binding transport proteins which bind Fe(3+) ion in association with the binding of an anion, usually bicarbonate.
Similarity
Belongs to the amiloride-sensitive sodium channel (TC 1.A.6) family.
Belongs to the transferrin family.
Belongs to the transferrin family.
Feature
chain Transferrin
Uniprot
H9JRR1
A0A067YAW9
A0A1E1WT36
A0A1E1WHK8
A0A194Q1N2
A0A2W1B0P0
+ More
A0A2H1VH25 A0A3S2LXA7 A0A2H1V887 A0A1Q3G4P5 A0A1Q3G4Q2 A0A1Q3G4N3 A0A182WF53 A0A340TBJ8 A0A182M421 A0A1J1IFB0 A0A084WBF0 Q7Q328 A0A182R8Q5 W5JTI5 T1DQ95 A0A182YPG1 A0A182FTD3 E2C2G6 A0A182MXW7 A0A182P9D2 A0A182IVG1 A0A182QXI1 A0A182KC18 A0A182VI64 A0A026WSE4 Q16PI3 A0A158NFY7 A0A195B271 A0A182HNK8 A0A195F0Y0 A0A182UGP9 A0A195E7T3 E9J634 A0A0C9R5E0 E2AMH2 A0A232EN02 A0A336M1Q9 K7JA32 A0A336LPD1 A0A151IQC0 A0A1B6HWX7 A0A2J7RCI4 F4X6X9 A0A336MCS2 A0A336ML09 A0A151XBZ1 A0A2P8Z1N8 A0A1B6GCI2 A0A1V1FVP3 A0A1Y1K651 A0A1Y1K3F2 A0A1Y1K3F3 A0A1Y1K3F6 A0A1B0CUY5 A0A182LPI1 A0A2J7RCI5 A0A1L8DIX9 A0A1L8DJ83 A0A1L8DIY4 A0A1L8DIZ1 A0A0P4VVZ2 V5I8N4 A0A1W4WK07 A0A1W4WVU4 A0A0A9XS57 A0A0K8T0U4 A0A224XKM5 A0A2J7RCI6 A0A1B0GHL1 A0A139WNP2 A0A139WNP6 T1HAU6 A0A0T6B7B3 A0A067R8G2 A0A2H4N2F8 A0A2J7RCH3 Q8MU80 A0A2J7RCG2 E9G2A9 A0A139WP46
A0A2H1VH25 A0A3S2LXA7 A0A2H1V887 A0A1Q3G4P5 A0A1Q3G4Q2 A0A1Q3G4N3 A0A182WF53 A0A340TBJ8 A0A182M421 A0A1J1IFB0 A0A084WBF0 Q7Q328 A0A182R8Q5 W5JTI5 T1DQ95 A0A182YPG1 A0A182FTD3 E2C2G6 A0A182MXW7 A0A182P9D2 A0A182IVG1 A0A182QXI1 A0A182KC18 A0A182VI64 A0A026WSE4 Q16PI3 A0A158NFY7 A0A195B271 A0A182HNK8 A0A195F0Y0 A0A182UGP9 A0A195E7T3 E9J634 A0A0C9R5E0 E2AMH2 A0A232EN02 A0A336M1Q9 K7JA32 A0A336LPD1 A0A151IQC0 A0A1B6HWX7 A0A2J7RCI4 F4X6X9 A0A336MCS2 A0A336ML09 A0A151XBZ1 A0A2P8Z1N8 A0A1B6GCI2 A0A1V1FVP3 A0A1Y1K651 A0A1Y1K3F2 A0A1Y1K3F3 A0A1Y1K3F6 A0A1B0CUY5 A0A182LPI1 A0A2J7RCI5 A0A1L8DIX9 A0A1L8DJ83 A0A1L8DIY4 A0A1L8DIZ1 A0A0P4VVZ2 V5I8N4 A0A1W4WK07 A0A1W4WVU4 A0A0A9XS57 A0A0K8T0U4 A0A224XKM5 A0A2J7RCI6 A0A1B0GHL1 A0A139WNP2 A0A139WNP6 T1HAU6 A0A0T6B7B3 A0A067R8G2 A0A2H4N2F8 A0A2J7RCH3 Q8MU80 A0A2J7RCG2 E9G2A9 A0A139WP46
Pubmed
EMBL
BABH01040835
BABH01040836
KC456662
AGZ62936.1
GDQN01000894
JAT90160.1
+ More
GDQN01004606 JAT86448.1 KQ459580 KPI99233.1 KZ150499 PZC70669.1 ODYU01002337 SOQ39702.1 RSAL01000132 RVE46379.1 ODYU01001191 SOQ37063.1 GFDL01000282 JAV34763.1 GFDL01000267 JAV34778.1 GFDL01000284 JAV34761.1 AXCM01000254 CVRI01000047 CRK97686.1 ATLV01022347 KE525331 KFB47544.1 AAAB01008966 EAA12967.4 ADMH02000540 ETN66054.1 GAMD01001295 JAB00296.1 GL452135 EFN77822.1 AXCN02002114 KK107135 QOIP01000006 EZA58009.1 RLU21675.1 CH477781 EAT36265.1 ADTU01014841 KQ976662 KYM78568.1 APCN01001939 KQ981864 KYN34235.1 KQ979568 KYN20902.1 GL768221 EFZ11734.1 GBYB01011524 JAG81291.1 GL440813 EFN65396.1 NNAY01003269 OXU19707.1 UFQS01000099 UFQT01000099 SSW99518.1 SSX19898.1 AAZX01005059 AAZX01016347 SSW99516.1 SSX19896.1 KQ976780 KYN08345.1 GECU01028543 JAS79163.1 NEVH01005885 PNF38540.1 GL888818 EGI57911.1 UFQT01000715 SSX26709.1 SSX26708.1 KQ982314 KYQ57891.1 PYGN01000239 PSN50413.1 GECZ01009630 JAS60139.1 FX985455 BAX07468.1 GEZM01094052 JAV55971.1 GEZM01094051 JAV55972.1 GEZM01094049 JAV55974.1 GEZM01094050 JAV55973.1 AJWK01029837 PNF38542.1 GFDF01007655 JAV06429.1 GFDF01007657 JAV06427.1 GFDF01007656 JAV06428.1 GFDF01007658 JAV06426.1 GDKW01000354 JAI56241.1 GALX01004380 JAB64086.1 GBHO01021123 GDHC01021803 JAG22481.1 JAP96825.1 GBRD01006692 JAG59129.1 GFTR01007857 JAW08569.1 PNF38541.1 AJWK01005316 KQ971308 KYB29544.1 KYB29543.1 ACPB03017612 LJIG01009362 KRT83232.1 KK852636 KDR19744.1 KY653081 ATU47275.1 PNF38525.1 AF535146 AAN03488.1 PNF38524.1 GL732530 EFX86336.1 KYB29545.1
GDQN01004606 JAT86448.1 KQ459580 KPI99233.1 KZ150499 PZC70669.1 ODYU01002337 SOQ39702.1 RSAL01000132 RVE46379.1 ODYU01001191 SOQ37063.1 GFDL01000282 JAV34763.1 GFDL01000267 JAV34778.1 GFDL01000284 JAV34761.1 AXCM01000254 CVRI01000047 CRK97686.1 ATLV01022347 KE525331 KFB47544.1 AAAB01008966 EAA12967.4 ADMH02000540 ETN66054.1 GAMD01001295 JAB00296.1 GL452135 EFN77822.1 AXCN02002114 KK107135 QOIP01000006 EZA58009.1 RLU21675.1 CH477781 EAT36265.1 ADTU01014841 KQ976662 KYM78568.1 APCN01001939 KQ981864 KYN34235.1 KQ979568 KYN20902.1 GL768221 EFZ11734.1 GBYB01011524 JAG81291.1 GL440813 EFN65396.1 NNAY01003269 OXU19707.1 UFQS01000099 UFQT01000099 SSW99518.1 SSX19898.1 AAZX01005059 AAZX01016347 SSW99516.1 SSX19896.1 KQ976780 KYN08345.1 GECU01028543 JAS79163.1 NEVH01005885 PNF38540.1 GL888818 EGI57911.1 UFQT01000715 SSX26709.1 SSX26708.1 KQ982314 KYQ57891.1 PYGN01000239 PSN50413.1 GECZ01009630 JAS60139.1 FX985455 BAX07468.1 GEZM01094052 JAV55971.1 GEZM01094051 JAV55972.1 GEZM01094049 JAV55974.1 GEZM01094050 JAV55973.1 AJWK01029837 PNF38542.1 GFDF01007655 JAV06429.1 GFDF01007657 JAV06427.1 GFDF01007656 JAV06428.1 GFDF01007658 JAV06426.1 GDKW01000354 JAI56241.1 GALX01004380 JAB64086.1 GBHO01021123 GDHC01021803 JAG22481.1 JAP96825.1 GBRD01006692 JAG59129.1 GFTR01007857 JAW08569.1 PNF38541.1 AJWK01005316 KQ971308 KYB29544.1 KYB29543.1 ACPB03017612 LJIG01009362 KRT83232.1 KK852636 KDR19744.1 KY653081 ATU47275.1 PNF38525.1 AF535146 AAN03488.1 PNF38524.1 GL732530 EFX86336.1 KYB29545.1
Proteomes
UP000005204
UP000053268
UP000283053
UP000075920
UP000076407
UP000075883
+ More
UP000183832 UP000030765 UP000007062 UP000075900 UP000000673 UP000076408 UP000069272 UP000008237 UP000075884 UP000075885 UP000075880 UP000075886 UP000075881 UP000075903 UP000053097 UP000279307 UP000008820 UP000005205 UP000078540 UP000075840 UP000078541 UP000075902 UP000078492 UP000000311 UP000215335 UP000002358 UP000078542 UP000235965 UP000007755 UP000075809 UP000245037 UP000092461 UP000075882 UP000192223 UP000007266 UP000015103 UP000027135 UP000000305
UP000183832 UP000030765 UP000007062 UP000075900 UP000000673 UP000076408 UP000069272 UP000008237 UP000075884 UP000075885 UP000075880 UP000075886 UP000075881 UP000075903 UP000053097 UP000279307 UP000008820 UP000005205 UP000078540 UP000075840 UP000078541 UP000075902 UP000078492 UP000000311 UP000215335 UP000002358 UP000078542 UP000235965 UP000007755 UP000075809 UP000245037 UP000092461 UP000075882 UP000192223 UP000007266 UP000015103 UP000027135 UP000000305
PRIDE
Pfam
Interpro
IPR001156
Transferrin-like_dom
+ More
IPR001873 ENaC
IPR016357 Transferrin
IPR034223 IGCP-like_CPD
IPR000834 Peptidase_M14
IPR029249 Rotatin_N
IPR030791 Rotatin
IPR006561 DZF_dom
IPR006116 2-5-oligoadenylate_synth_N
IPR000690 Matrin/U1-C_Znf_C2H2
IPR031774 SF3A3_dom
IPR024598 DUF3449
IPR031776 SF3A3
IPR021966 SF3a60_bindingd
IPR018195 Transferrin_Fe_BS
IPR001873 ENaC
IPR016357 Transferrin
IPR034223 IGCP-like_CPD
IPR000834 Peptidase_M14
IPR029249 Rotatin_N
IPR030791 Rotatin
IPR006561 DZF_dom
IPR006116 2-5-oligoadenylate_synth_N
IPR000690 Matrin/U1-C_Znf_C2H2
IPR031774 SF3A3_dom
IPR024598 DUF3449
IPR031776 SF3A3
IPR021966 SF3a60_bindingd
IPR018195 Transferrin_Fe_BS
ProteinModelPortal
H9JRR1
A0A067YAW9
A0A1E1WT36
A0A1E1WHK8
A0A194Q1N2
A0A2W1B0P0
+ More
A0A2H1VH25 A0A3S2LXA7 A0A2H1V887 A0A1Q3G4P5 A0A1Q3G4Q2 A0A1Q3G4N3 A0A182WF53 A0A340TBJ8 A0A182M421 A0A1J1IFB0 A0A084WBF0 Q7Q328 A0A182R8Q5 W5JTI5 T1DQ95 A0A182YPG1 A0A182FTD3 E2C2G6 A0A182MXW7 A0A182P9D2 A0A182IVG1 A0A182QXI1 A0A182KC18 A0A182VI64 A0A026WSE4 Q16PI3 A0A158NFY7 A0A195B271 A0A182HNK8 A0A195F0Y0 A0A182UGP9 A0A195E7T3 E9J634 A0A0C9R5E0 E2AMH2 A0A232EN02 A0A336M1Q9 K7JA32 A0A336LPD1 A0A151IQC0 A0A1B6HWX7 A0A2J7RCI4 F4X6X9 A0A336MCS2 A0A336ML09 A0A151XBZ1 A0A2P8Z1N8 A0A1B6GCI2 A0A1V1FVP3 A0A1Y1K651 A0A1Y1K3F2 A0A1Y1K3F3 A0A1Y1K3F6 A0A1B0CUY5 A0A182LPI1 A0A2J7RCI5 A0A1L8DIX9 A0A1L8DJ83 A0A1L8DIY4 A0A1L8DIZ1 A0A0P4VVZ2 V5I8N4 A0A1W4WK07 A0A1W4WVU4 A0A0A9XS57 A0A0K8T0U4 A0A224XKM5 A0A2J7RCI6 A0A1B0GHL1 A0A139WNP2 A0A139WNP6 T1HAU6 A0A0T6B7B3 A0A067R8G2 A0A2H4N2F8 A0A2J7RCH3 Q8MU80 A0A2J7RCG2 E9G2A9 A0A139WP46
A0A2H1VH25 A0A3S2LXA7 A0A2H1V887 A0A1Q3G4P5 A0A1Q3G4Q2 A0A1Q3G4N3 A0A182WF53 A0A340TBJ8 A0A182M421 A0A1J1IFB0 A0A084WBF0 Q7Q328 A0A182R8Q5 W5JTI5 T1DQ95 A0A182YPG1 A0A182FTD3 E2C2G6 A0A182MXW7 A0A182P9D2 A0A182IVG1 A0A182QXI1 A0A182KC18 A0A182VI64 A0A026WSE4 Q16PI3 A0A158NFY7 A0A195B271 A0A182HNK8 A0A195F0Y0 A0A182UGP9 A0A195E7T3 E9J634 A0A0C9R5E0 E2AMH2 A0A232EN02 A0A336M1Q9 K7JA32 A0A336LPD1 A0A151IQC0 A0A1B6HWX7 A0A2J7RCI4 F4X6X9 A0A336MCS2 A0A336ML09 A0A151XBZ1 A0A2P8Z1N8 A0A1B6GCI2 A0A1V1FVP3 A0A1Y1K651 A0A1Y1K3F2 A0A1Y1K3F3 A0A1Y1K3F6 A0A1B0CUY5 A0A182LPI1 A0A2J7RCI5 A0A1L8DIX9 A0A1L8DJ83 A0A1L8DIY4 A0A1L8DIZ1 A0A0P4VVZ2 V5I8N4 A0A1W4WK07 A0A1W4WVU4 A0A0A9XS57 A0A0K8T0U4 A0A224XKM5 A0A2J7RCI6 A0A1B0GHL1 A0A139WNP2 A0A139WNP6 T1HAU6 A0A0T6B7B3 A0A067R8G2 A0A2H4N2F8 A0A2J7RCH3 Q8MU80 A0A2J7RCG2 E9G2A9 A0A139WP46
PDB
1JW1
E-value=1.64534e-17,
Score=222
Ontologies
GO
PANTHER
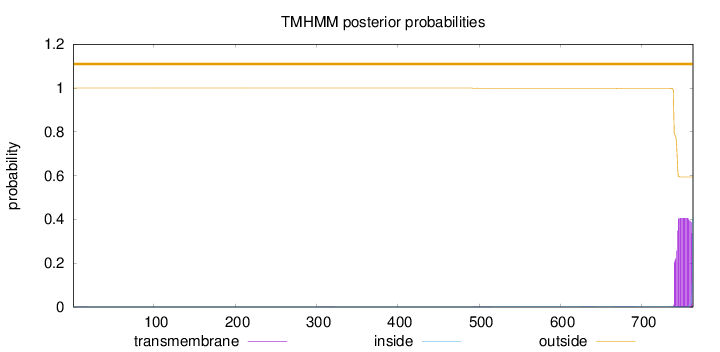
Topology
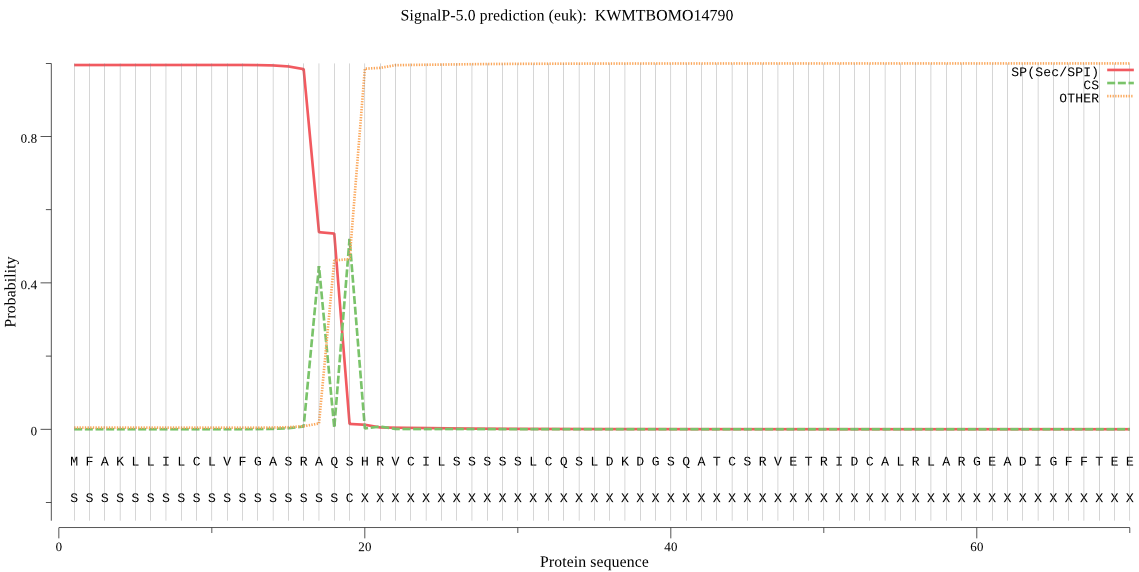
SignalP
Position: 1 - 19,
Likelihood: 0.995356
Length:
763
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
8.49233
Exp number, first 60 AAs:
0.00285
Total prob of N-in:
0.00032
outside
1 - 763
Population Genetic Test Statistics
Pi
221.667529
Theta
151.769522
Tajima's D
1.548414
CLR
0.184594
CSRT
0.797910104494775
Interpretation
Uncertain
