Gene
KWMTBOMO14788 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012217
Annotation
hemolymph_protein_14_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 4.103
Sequence
CDS
ATGTCCAGATGCCAGAGCAATTGGTTCCGCTGTACGTATGGCGCGTGCGTGGATGGTACGGCGCCCTGCAACGGAATCCAGGAGTGCGCCGATAACTCTGATGAGCTGCTACCTCAGTGTCGGGGGAATGAGGTCGCGGACGCGACGAACTTGAGAGACAAGTTCAAGTGTGCTGACGGTAACGTTATACCGGCCACCGCGTTCTGTGACGGCGTAGCCGACTGCCCGGACGGGTCTGACGAGACGGTCCAGGCGTGCGCGGGCAGGACGTGCCCGGCGTACTTGTTCCAGTGCGCGTATGGAGCTTGCGTAGACGCCGGATCAGACTGTAACGGGATAAAAGAATGCGCTGACGGCTCGGATGAGTCGGACGAGCTCTGTGATAGAGTCCTGTCCGGGAAGCCGACGCAAAAGCCTTCAGCATCAGGCGGATGCGTCCTGCCCCAGTACCCAGCTCACGGCTCGTACGTGGTCCTGAACACGCCGAACGCTACTCCCGGCCAGACCTTCGACTCGATCCAAGTGAACGTGACCTGCAAGCCGGGGTACGGCGTGATGGGCAGGAACGACGTCTTCTGCTTGAACGGGTGGTGGTCGGACAAGCTGCCTCAGTGCGTCCGTTTCTGCACGCTCAACAGTCATGCTAGCGTCGAGTACAAATGTCTGCTCACCAATGGTAACGAGGGTACCAGACCCTGCGGCTCCCTGGAGCCGACCGGCACAATAGTCCAGCCGCAGTGCAGGTCCCCCAACTATTACTCGACCGTGCCGCTCGGCTACATGCGGTGCATCGAGGGGAACTGGGACTACGTGGCCACATGCACTCCAGAATGCGGCAGAGTCACGCCTGACGGTGAAGAGTTGGTGTTCGGCGGGAGATCAGCGAAAAGAGGAGAACTACCCTGGCATGCTGGCATATATAGGAAGACTAGCACACCGTACATGCAGATCTGCGGTGGATCTCTCGTTAGCACCACCGTCGTCATATCAGCCGCTCACTGCTTCTGGAGCAACCTCAACCAGCAGCTACCCACAACTGACTATGCCGTTGCCGTCGGTAAGATTTACAGACCCTGGAACAATCCGAAGGACTTGGACGTCCAGAAATCGGATGTGAGGGATATAAAAATCCCGATAAGATTCCAAGGGAGGACGGCGAACTTTCAAGAGGACATAGCGATCGTGATACTGGCGACAAGGATCGAGTACCGCACCTACGTCAGGCCGGCCTGTCTGGACTTCGACATCAGCTTTGAGAGGCAGCAACTACAAGACGGGAAACCGGGCAAGGTGGCCGGTTGGGGTTTGACTTCGGAAAACGGCAACGCCTCGCAAATACTGAAAGTCGTTGATTTGCCATACATCGACATAGATAGGTGCATCGGCATGGCGCCTCCCGGGTTCAGGGAGTACATTAATGGCGACAAGATCTGCGCTGGCGACGGAGAAGGCAAAGCTTTATGCAAAGGGGACAGCGGCGGGGGCCTAGCTTTTTCCACTTACGACAAGGGAGTCGAAAGATACTATTTACGCGGAATCGTGTCAACGGCTTCGACGAACGAGAATCTGTGCAACACAAACACATTCACAACATTCACACAAGTCTCGAAACACGAACATTTCATCAAGCAATATCTGCACGTGGTTCAGACTTAG
Protein
MSRCQSNWFRCTYGACVDGTAPCNGIQECADNSDELLPQCRGNEVADATNLRDKFKCADGNVIPATAFCDGVADCPDGSDETVQACAGRTCPAYLFQCAYGACVDAGSDCNGIKECADGSDESDELCDRVLSGKPTQKPSASGGCVLPQYPAHGSYVVLNTPNATPGQTFDSIQVNVTCKPGYGVMGRNDVFCLNGWWSDKLPQCVRFCTLNSHASVEYKCLLTNGNEGTRPCGSLEPTGTIVQPQCRSPNYYSTVPLGYMRCIEGNWDYVATCTPECGRVTPDGEELVFGGRSAKRGELPWHAGIYRKTSTPYMQICGGSLVSTTVVISAAHCFWSNLNQQLPTTDYAVAVGKIYRPWNNPKDLDVQKSDVRDIKIPIRFQGRTANFQEDIAIVILATRIEYRTYVRPACLDFDISFERQQLQDGKPGKVAGWGLTSENGNASQILKVVDLPYIDIDRCIGMAPPGFREYINGDKICAGDGEGKALCKGDSGGGLAFSTYDKGVERYYLRGIVSTASTNENLCNTNTFTTFTQVSKHEHFIKQYLHVVQT
Summary
Similarity
Belongs to the peptidase S1 family.
Belongs to the mitochondrial carrier (TC 2.A.29) family.
Belongs to the mitochondrial carrier (TC 2.A.29) family.
Uniprot
I3WAD0
A0A2H1VKP2
Q69BL0
A0A2W1BIK5
A0A2A4J653
A0A2A4J8A8
+ More
A0A2A4JXS9 Q5MGG8 A0A194Q760 A0A0N1PIR0 A0A2W1BGN3 A0A0N1PGE4 A0A194Q0Y4 A0A0N1I6Q2 A0A3S2LXN3 A0A2H1VLP9 A0A0N1PIX0 A0A2P8YP29 A0A0L7LGL1 A0A2J7RCH1 A0A067RJX7 A0A182LSY4 A0A182Q107 A0A0N1I8Z7 A0A182VRJ4 U5EPN9 A0A1Q3FMR3 A0A182GDV9 Q17H79 B0X3L4 A0A2M4AGF6 A0A2M4AFU1 A0A182NFK7 W5JJ60 A0A182X2R5 A0A182H7R9 A0A182HGT4 A0A182F8Q3 A0A182ITD4 A0A1S4GDQ4 Q7PY92 A0A2M4BGF8 A0A182KMB4 A0A2M4BGG7 A0A182RNY9 A0A0A9YJE5 A0A146LVV0 A0A224X923 A0A182TRW0 A0A182VJ48 T1H816 A0A2K8JMG0 D6WBK5 A0A1W4XIN2 A0A1W4XTP2 B1B5K3 A0A182XV86 A0A1Y1LR53 D6WAZ7 A0A170ZNF0 A0A1Y1LR62 A0A182P9S2 A0A084VWP2 A0A2A4J979 A0A1V1FIV2 J9JRX5 A0A154PEE3 A0A1L8DQQ0 A0A1J1J0Z5 A0A310SF19 N6USS0 U4UFJ4 A0A2A3ER96 A0A1B6E5L3 A0A088AKU8 J9K1E4 A0A0L7RG32 A0A1A9ZTA3 A0A1A9V5K8 A0A146LPF0 A0A1B0FPU9
A0A2A4JXS9 Q5MGG8 A0A194Q760 A0A0N1PIR0 A0A2W1BGN3 A0A0N1PGE4 A0A194Q0Y4 A0A0N1I6Q2 A0A3S2LXN3 A0A2H1VLP9 A0A0N1PIX0 A0A2P8YP29 A0A0L7LGL1 A0A2J7RCH1 A0A067RJX7 A0A182LSY4 A0A182Q107 A0A0N1I8Z7 A0A182VRJ4 U5EPN9 A0A1Q3FMR3 A0A182GDV9 Q17H79 B0X3L4 A0A2M4AGF6 A0A2M4AFU1 A0A182NFK7 W5JJ60 A0A182X2R5 A0A182H7R9 A0A182HGT4 A0A182F8Q3 A0A182ITD4 A0A1S4GDQ4 Q7PY92 A0A2M4BGF8 A0A182KMB4 A0A2M4BGG7 A0A182RNY9 A0A0A9YJE5 A0A146LVV0 A0A224X923 A0A182TRW0 A0A182VJ48 T1H816 A0A2K8JMG0 D6WBK5 A0A1W4XIN2 A0A1W4XTP2 B1B5K3 A0A182XV86 A0A1Y1LR53 D6WAZ7 A0A170ZNF0 A0A1Y1LR62 A0A182P9S2 A0A084VWP2 A0A2A4J979 A0A1V1FIV2 J9JRX5 A0A154PEE3 A0A1L8DQQ0 A0A1J1J0Z5 A0A310SF19 N6USS0 U4UFJ4 A0A2A3ER96 A0A1B6E5L3 A0A088AKU8 J9K1E4 A0A0L7RG32 A0A1A9ZTA3 A0A1A9V5K8 A0A146LPF0 A0A1B0FPU9
Pubmed
EMBL
JQ954757
AFK93534.1
ODYU01002842
SOQ40824.1
AY380790
AAR29602.1
+ More
KZ150174 PZC72580.1 NWSH01003130 PCG66903.1 NWSH01002430 PCG68315.1 NWSH01000454 PCG76303.1 AY829818 AAV91432.2 KQ459580 KPI99230.1 KQ460246 KPJ16373.1 KZ150237 PZC71960.1 KQ460735 KPJ12635.1 KPI99211.1 KPJ12638.1 RSAL01000132 RVE46370.1 ODYU01003240 SOQ41738.1 KPJ12637.1 PYGN01000456 PSN46002.1 JTDY01001173 KOB74658.1 NEVH01005885 PNF38537.1 KK852636 KDR19731.1 AXCM01003686 AXCN02000208 KPJ16374.1 GANO01004615 JAB55256.1 GFDL01006237 JAV28808.1 JXUM01056539 JXUM01056540 KQ561915 KXJ77171.1 CH477252 EAT46012.1 DS232317 EDS39906.1 GGFK01006544 MBW39865.1 GGFK01006333 MBW39654.1 ADMH02001351 ETN62915.1 JXUM01028537 KQ560824 KXJ80737.1 APCN01005945 AAAB01008987 EAA01121.5 GGFJ01003008 MBW52149.1 GGFJ01003009 MBW52150.1 GBHO01011848 GBHO01011846 GBRD01011126 JAG31756.1 JAG31758.1 JAG54698.1 GDHC01007812 JAQ10817.1 GFTR01007569 JAW08857.1 ACPB03011663 KY031267 ATU83018.1 KQ971311 EEZ99332.2 AB363982 BAG14264.1 GEZM01049076 JAV76122.1 EEZ98916.1 GEMB01002039 JAS01140.1 GEZM01049075 JAV76123.1 ATLV01017677 ATLV01017678 ATLV01017679 KE525181 KFB42386.1 PCG68316.1 FX985348 BAX07361.1 ABLF02017553 KQ434887 KZC10211.1 GFDF01005380 JAV08704.1 CVRI01000063 CRL04521.1 KQ765323 KQ759877 OAD53988.1 OAD62273.1 APGK01017806 APGK01017807 APGK01017808 KB740053 ENN81797.1 KB632093 ERL88690.1 KZ288195 PBC33792.1 GEDC01004111 JAS33187.1 ABLF02038724 ABLF02038736 KQ414599 KOC69793.1 GDHC01009430 JAQ09199.1 CCAG010010748 CCAG010010749 CCAG010010750 CCAG010010751
KZ150174 PZC72580.1 NWSH01003130 PCG66903.1 NWSH01002430 PCG68315.1 NWSH01000454 PCG76303.1 AY829818 AAV91432.2 KQ459580 KPI99230.1 KQ460246 KPJ16373.1 KZ150237 PZC71960.1 KQ460735 KPJ12635.1 KPI99211.1 KPJ12638.1 RSAL01000132 RVE46370.1 ODYU01003240 SOQ41738.1 KPJ12637.1 PYGN01000456 PSN46002.1 JTDY01001173 KOB74658.1 NEVH01005885 PNF38537.1 KK852636 KDR19731.1 AXCM01003686 AXCN02000208 KPJ16374.1 GANO01004615 JAB55256.1 GFDL01006237 JAV28808.1 JXUM01056539 JXUM01056540 KQ561915 KXJ77171.1 CH477252 EAT46012.1 DS232317 EDS39906.1 GGFK01006544 MBW39865.1 GGFK01006333 MBW39654.1 ADMH02001351 ETN62915.1 JXUM01028537 KQ560824 KXJ80737.1 APCN01005945 AAAB01008987 EAA01121.5 GGFJ01003008 MBW52149.1 GGFJ01003009 MBW52150.1 GBHO01011848 GBHO01011846 GBRD01011126 JAG31756.1 JAG31758.1 JAG54698.1 GDHC01007812 JAQ10817.1 GFTR01007569 JAW08857.1 ACPB03011663 KY031267 ATU83018.1 KQ971311 EEZ99332.2 AB363982 BAG14264.1 GEZM01049076 JAV76122.1 EEZ98916.1 GEMB01002039 JAS01140.1 GEZM01049075 JAV76123.1 ATLV01017677 ATLV01017678 ATLV01017679 KE525181 KFB42386.1 PCG68316.1 FX985348 BAX07361.1 ABLF02017553 KQ434887 KZC10211.1 GFDF01005380 JAV08704.1 CVRI01000063 CRL04521.1 KQ765323 KQ759877 OAD53988.1 OAD62273.1 APGK01017806 APGK01017807 APGK01017808 KB740053 ENN81797.1 KB632093 ERL88690.1 KZ288195 PBC33792.1 GEDC01004111 JAS33187.1 ABLF02038724 ABLF02038736 KQ414599 KOC69793.1 GDHC01009430 JAQ09199.1 CCAG010010748 CCAG010010749 CCAG010010750 CCAG010010751
Proteomes
UP000218220
UP000053268
UP000053240
UP000283053
UP000245037
UP000037510
+ More
UP000235965 UP000027135 UP000075883 UP000075886 UP000075920 UP000069940 UP000249989 UP000008820 UP000002320 UP000075884 UP000000673 UP000076407 UP000075840 UP000069272 UP000075880 UP000007062 UP000075882 UP000075900 UP000075902 UP000075903 UP000015103 UP000007266 UP000192223 UP000076408 UP000075885 UP000030765 UP000007819 UP000076502 UP000183832 UP000019118 UP000030742 UP000242457 UP000005203 UP000053825 UP000092445 UP000078200 UP000092444
UP000235965 UP000027135 UP000075883 UP000075886 UP000075920 UP000069940 UP000249989 UP000008820 UP000002320 UP000075884 UP000000673 UP000076407 UP000075840 UP000069272 UP000075880 UP000007062 UP000075882 UP000075900 UP000075902 UP000075903 UP000015103 UP000007266 UP000192223 UP000076408 UP000075885 UP000030765 UP000007819 UP000076502 UP000183832 UP000019118 UP000030742 UP000242457 UP000005203 UP000053825 UP000092445 UP000078200 UP000092444
PRIDE
Pfam
Interpro
IPR023415
LDLR_class-A_CS
+ More
IPR000436 Sushi_SCR_CCP_dom
IPR018114 TRYPSIN_HIS
IPR001254 Trypsin_dom
IPR009003 Peptidase_S1_PA
IPR002172 LDrepeatLR_classA_rpt
IPR036055 LDL_receptor-like_sf
IPR035976 Sushi/SCR/CCP_sf
IPR001314 Peptidase_S1A
IPR018108 Mitochondrial_sb/sol_carrier
IPR023395 Mt_carrier_dom_sf
IPR013032 EGF-like_CS
IPR000742 EGF-like_dom
IPR001879 GPCR_2_extracellular_dom
IPR003593 AAA+_ATPase
IPR001487 Bromodomain
IPR020095 PsdUridine_synth_TruA_C
IPR003959 ATPase_AAA_core
IPR020103 PsdUridine_synth_cat_dom_sf
IPR036427 Bromodomain-like_sf
IPR001406 PsdUridine_synth_TruA
IPR020097 PsdUridine_synth_TruA_a/b_dom
IPR041708 PUS1/PUS2-like
IPR027417 P-loop_NTPase
IPR003960 ATPase_AAA_CS
IPR033116 TRYPSIN_SER
IPR000436 Sushi_SCR_CCP_dom
IPR018114 TRYPSIN_HIS
IPR001254 Trypsin_dom
IPR009003 Peptidase_S1_PA
IPR002172 LDrepeatLR_classA_rpt
IPR036055 LDL_receptor-like_sf
IPR035976 Sushi/SCR/CCP_sf
IPR001314 Peptidase_S1A
IPR018108 Mitochondrial_sb/sol_carrier
IPR023395 Mt_carrier_dom_sf
IPR013032 EGF-like_CS
IPR000742 EGF-like_dom
IPR001879 GPCR_2_extracellular_dom
IPR003593 AAA+_ATPase
IPR001487 Bromodomain
IPR020095 PsdUridine_synth_TruA_C
IPR003959 ATPase_AAA_core
IPR020103 PsdUridine_synth_cat_dom_sf
IPR036427 Bromodomain-like_sf
IPR001406 PsdUridine_synth_TruA
IPR020097 PsdUridine_synth_TruA_a/b_dom
IPR041708 PUS1/PUS2-like
IPR027417 P-loop_NTPase
IPR003960 ATPase_AAA_CS
IPR033116 TRYPSIN_SER
SUPFAM
Gene 3D
ProteinModelPortal
I3WAD0
A0A2H1VKP2
Q69BL0
A0A2W1BIK5
A0A2A4J653
A0A2A4J8A8
+ More
A0A2A4JXS9 Q5MGG8 A0A194Q760 A0A0N1PIR0 A0A2W1BGN3 A0A0N1PGE4 A0A194Q0Y4 A0A0N1I6Q2 A0A3S2LXN3 A0A2H1VLP9 A0A0N1PIX0 A0A2P8YP29 A0A0L7LGL1 A0A2J7RCH1 A0A067RJX7 A0A182LSY4 A0A182Q107 A0A0N1I8Z7 A0A182VRJ4 U5EPN9 A0A1Q3FMR3 A0A182GDV9 Q17H79 B0X3L4 A0A2M4AGF6 A0A2M4AFU1 A0A182NFK7 W5JJ60 A0A182X2R5 A0A182H7R9 A0A182HGT4 A0A182F8Q3 A0A182ITD4 A0A1S4GDQ4 Q7PY92 A0A2M4BGF8 A0A182KMB4 A0A2M4BGG7 A0A182RNY9 A0A0A9YJE5 A0A146LVV0 A0A224X923 A0A182TRW0 A0A182VJ48 T1H816 A0A2K8JMG0 D6WBK5 A0A1W4XIN2 A0A1W4XTP2 B1B5K3 A0A182XV86 A0A1Y1LR53 D6WAZ7 A0A170ZNF0 A0A1Y1LR62 A0A182P9S2 A0A084VWP2 A0A2A4J979 A0A1V1FIV2 J9JRX5 A0A154PEE3 A0A1L8DQQ0 A0A1J1J0Z5 A0A310SF19 N6USS0 U4UFJ4 A0A2A3ER96 A0A1B6E5L3 A0A088AKU8 J9K1E4 A0A0L7RG32 A0A1A9ZTA3 A0A1A9V5K8 A0A146LPF0 A0A1B0FPU9
A0A2A4JXS9 Q5MGG8 A0A194Q760 A0A0N1PIR0 A0A2W1BGN3 A0A0N1PGE4 A0A194Q0Y4 A0A0N1I6Q2 A0A3S2LXN3 A0A2H1VLP9 A0A0N1PIX0 A0A2P8YP29 A0A0L7LGL1 A0A2J7RCH1 A0A067RJX7 A0A182LSY4 A0A182Q107 A0A0N1I8Z7 A0A182VRJ4 U5EPN9 A0A1Q3FMR3 A0A182GDV9 Q17H79 B0X3L4 A0A2M4AGF6 A0A2M4AFU1 A0A182NFK7 W5JJ60 A0A182X2R5 A0A182H7R9 A0A182HGT4 A0A182F8Q3 A0A182ITD4 A0A1S4GDQ4 Q7PY92 A0A2M4BGF8 A0A182KMB4 A0A2M4BGG7 A0A182RNY9 A0A0A9YJE5 A0A146LVV0 A0A224X923 A0A182TRW0 A0A182VJ48 T1H816 A0A2K8JMG0 D6WBK5 A0A1W4XIN2 A0A1W4XTP2 B1B5K3 A0A182XV86 A0A1Y1LR53 D6WAZ7 A0A170ZNF0 A0A1Y1LR62 A0A182P9S2 A0A084VWP2 A0A2A4J979 A0A1V1FIV2 J9JRX5 A0A154PEE3 A0A1L8DQQ0 A0A1J1J0Z5 A0A310SF19 N6USS0 U4UFJ4 A0A2A3ER96 A0A1B6E5L3 A0A088AKU8 J9K1E4 A0A0L7RG32 A0A1A9ZTA3 A0A1A9V5K8 A0A146LPF0 A0A1B0FPU9
PDB
4IGD
E-value=2.25354e-22,
Score=262
Ontologies
KEGG
GO
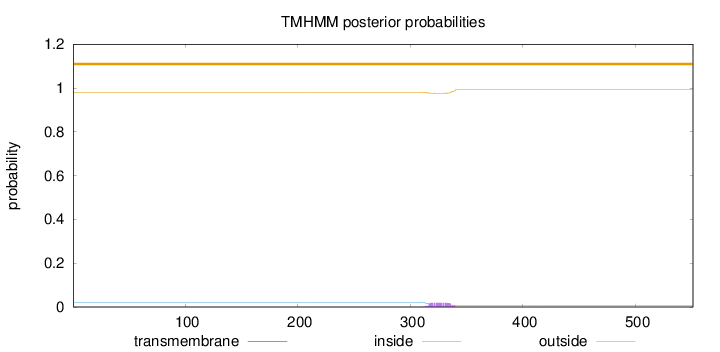
Topology
Length:
551
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.4177
Exp number, first 60 AAs:
0.0002
Total prob of N-in:
0.01991
outside
1 - 551
Population Genetic Test Statistics
Pi
248.187166
Theta
154.522514
Tajima's D
1.804845
CLR
0
CSRT
0.846857657117144
Interpretation
Uncertain
