Gene
KWMTBOMO14787
Annotation
hemolymph_protein_14_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.719
Sequence
CDS
ATGTATTTATATTTATCGGTAATATTGTTCGTAACGTCTTCTGTCCTGACGAAGATCACCAATCCATACGACAGCTCGTGCAGCTACAACGAGTTTGAATGCGGAGATGGGGGCTGCGTGAGCATCGACAGAGTCTGCAACGGCATAGAGGATTGTTCTGGTGCCGAAGATGAGACTGTCTGCTACGAAATACCCGCAAAATTTGGATTGCATAGCTCTAGACGGCGTAGACAGACTTCTAATTGCAAGAAGTCGGAGTGGCGTTGCCGCGACGGTACGTGCATCTCGTTCGACGGCAAGTGCGACGGCGTCGTCGACTGTCCGGATTCGAGCGACGAAACCCATGCGCTTTGCGGGAAGACCACGTGA
Protein
MYLYLSVILFVTSSVLTKITNPYDSSCSYNEFECGDGGCVSIDRVCNGIEDCSGAEDETVCYEIPAKFGLHSSRRRRQTSNCKKSEWRCRDGTCISFDGKCDGVVDCPDSSDETHALCGKTT
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
I3WAD0
H9JRQ5
A0A0N0PDF1
A0A2H1VKP2
Q69BL0
A0A194Q0X1
+ More
A0A212FFN5 A0A2W1BIK5 A0A2W1BIE4 A0A2A4J8A8 A0A1B6CKY8 A0A195E4R2 A0A0Q9WWR1 A0A1B6EDX6 A0A1B6DS91 A0A2H1W8T0 A0A195BZV6 A0A0L7LDM0 Q297J7 U4UFJ4 N6USS0 B4G4A8 A0A0A1XDP6 A0A0A1X5C7 A0A182LSY4 A0A151WH43 A0A034WK22 A0A1D2N4X8 A0A224YPF5 W5JJ60 A0A3S2M4C7 A0A0Q9WYR9 A0A0Q9WV89 A0A0Q9X050 A0A084VUR2 A0A131YML4 A0A0Q9WN73 A0A1B0F0M3 A0A3L8DHC7 W8B7J5 A0A0Q9WN66 A0A0Q9X076 A0A0Q9WPG5 A0A0Q9WNJ7 W8AMV9 W8AJ92 A0A131Z9Z9 A0A0L8FZG5 J9K8R8 A0A0B7ATX1 A0A2M4AFU1 A0A2M4AGF6 B4L1X8 A0A3P9A5H3 A0A034WJ53 A0A1Y1MSQ2 A0A1Y1MUH0 A0A182F8Q3 A0A0L8GCJ0 A0A0L8GC81 A0A2M3ZZN1 A0A158NND7 G8Z9Y7 A0A0A1X1K7 A0A3Q3KPP9 A0A084VWP2 A0A195FXZ6 A8TEF0 A0A131XPK7 F1LET3 F1KYC8 A0A2M4B8R2 A0A182QRH4 F1KZ21 A0A0K8VDH4 A0A2A3ER96 A0A0L8GRS2 A0A3Q0JGI2 A0A310SF19 A0A224YUS3 A0A023FEK7 A0A131YP00 A0A224Z0F1 A0A131Z9J2 A0A0K8VQ98 A0A131Z8F7 W5M8L7 A0A210PXP2 A0A0N0BH65 A0A194R369 A0A310SBQ4 A0A182ITD4 A0A194PH43 A0A182Q107 T1E8X0 V9IHL7 R7U896
A0A212FFN5 A0A2W1BIK5 A0A2W1BIE4 A0A2A4J8A8 A0A1B6CKY8 A0A195E4R2 A0A0Q9WWR1 A0A1B6EDX6 A0A1B6DS91 A0A2H1W8T0 A0A195BZV6 A0A0L7LDM0 Q297J7 U4UFJ4 N6USS0 B4G4A8 A0A0A1XDP6 A0A0A1X5C7 A0A182LSY4 A0A151WH43 A0A034WK22 A0A1D2N4X8 A0A224YPF5 W5JJ60 A0A3S2M4C7 A0A0Q9WYR9 A0A0Q9WV89 A0A0Q9X050 A0A084VUR2 A0A131YML4 A0A0Q9WN73 A0A1B0F0M3 A0A3L8DHC7 W8B7J5 A0A0Q9WN66 A0A0Q9X076 A0A0Q9WPG5 A0A0Q9WNJ7 W8AMV9 W8AJ92 A0A131Z9Z9 A0A0L8FZG5 J9K8R8 A0A0B7ATX1 A0A2M4AFU1 A0A2M4AGF6 B4L1X8 A0A3P9A5H3 A0A034WJ53 A0A1Y1MSQ2 A0A1Y1MUH0 A0A182F8Q3 A0A0L8GCJ0 A0A0L8GC81 A0A2M3ZZN1 A0A158NND7 G8Z9Y7 A0A0A1X1K7 A0A3Q3KPP9 A0A084VWP2 A0A195FXZ6 A8TEF0 A0A131XPK7 F1LET3 F1KYC8 A0A2M4B8R2 A0A182QRH4 F1KZ21 A0A0K8VDH4 A0A2A3ER96 A0A0L8GRS2 A0A3Q0JGI2 A0A310SF19 A0A224YUS3 A0A023FEK7 A0A131YP00 A0A224Z0F1 A0A131Z9J2 A0A0K8VQ98 A0A131Z8F7 W5M8L7 A0A210PXP2 A0A0N0BH65 A0A194R369 A0A310SBQ4 A0A182ITD4 A0A194PH43 A0A182Q107 T1E8X0 V9IHL7 R7U896
Pubmed
EMBL
JQ954757
AFK93534.1
BABH01040844
KQ460389
KPJ15425.1
ODYU01002842
+ More
SOQ40824.1 AY380790 AAR29602.1 KQ459580 KPI99232.1 AGBW02008795 OWR52542.1 KZ150174 PZC72580.1 KZ150316 PZC71443.1 NWSH01002430 PCG68315.1 GEDC01023161 JAS14137.1 KQ979685 KYN19844.1 CH933810 KRF94150.1 GEDC01001166 JAS36132.1 GEDC01008789 JAS28509.1 ODYU01006758 SOQ48894.1 KQ978457 KYM93875.1 JTDY01001535 KOB73623.1 CM000070 EAL28208.2 KB632093 ERL88690.1 APGK01017806 APGK01017807 APGK01017808 KB740053 ENN81797.1 CH479179 EDW25146.1 GBXI01014482 GBXI01005217 JAC99809.1 JAD09075.1 GBXI01008342 JAD05950.1 AXCM01003686 KQ983136 KYQ47144.1 GAKP01004275 JAC54677.1 LJIJ01000233 ODN00115.1 GFPF01008342 MAA19488.1 ADMH02001351 ETN62915.1 RSAL01000038 RVE51080.1 KRF94153.1 KRF94156.1 KRF94154.1 ATLV01016989 ATLV01016990 KE525139 KFB41706.1 GEDV01008732 JAP79825.1 KRF94152.1 AJVK01007778 AJVK01007779 AJVK01007780 QOIP01000008 RLU19726.1 GAMC01020746 JAB85809.1 KRF94157.1 KRF94149.1 KRF94155.1 KRF94151.1 GAMC01020747 JAB85808.1 GAMC01020748 JAB85807.1 GEDV01000917 JAP87640.1 KQ425015 KOF70121.1 ABLF02033685 ABLF02033689 ABLF02033690 HACG01037197 CEK84062.1 GGFK01006333 MBW39654.1 GGFK01006544 MBW39865.1 EDW07699.2 GAKP01004273 JAC54679.1 GEZM01023937 GEZM01023935 GEZM01023934 JAV88128.1 GEZM01023936 GEZM01023933 GEZM01023932 JAV88130.1 KQ422575 KOF74633.1 KOF74632.1 GGFK01000644 MBW33965.1 ADTU01021233 ADTU01021234 ADTU01021235 ADTU01021236 ADTU01021237 ADTU01021238 ADTU01021239 ADTU01021240 ADTU01021241 ADTU01021242 GU454802 ADK55596.1 GBXI01009541 JAD04751.1 ATLV01017677 ATLV01017678 ATLV01017679 KE525181 KFB42386.1 KQ981208 KYN44709.1 EU024890 ABW79798.1 GEFH01001080 JAP67501.1 JI181595 ADY48637.1 JI167980 ADY42882.1 GGFJ01000077 MBW49218.1 AXCN02001967 JI168330 ADY43125.1 GDHF01030583 GDHF01015320 JAI21731.1 JAI36994.1 KZ288195 PBC33792.1 KQ420694 KOF79592.1 KQ765323 KQ759877 OAD53988.1 OAD62273.1 GFPF01008343 MAA19489.1 GBBK01005163 JAC19319.1 GEDV01008731 JAP79826.1 GFPF01008344 MAA19490.1 GEDV01000919 JAP87638.1 GDHF01011599 JAI40715.1 GEDV01000918 JAP87639.1 AHAT01019149 AHAT01019150 AHAT01019151 NEDP02005415 OWF41229.1 KQ435760 KOX75591.1 KQ460953 KPJ10291.1 KQ773953 OAD52336.1 KQ459605 KPI92029.1 AXCN02000208 GAMD01001822 JAA99768.1 JR043982 AEY59824.1 AMQN01008945 KB304240 ELU02214.1
SOQ40824.1 AY380790 AAR29602.1 KQ459580 KPI99232.1 AGBW02008795 OWR52542.1 KZ150174 PZC72580.1 KZ150316 PZC71443.1 NWSH01002430 PCG68315.1 GEDC01023161 JAS14137.1 KQ979685 KYN19844.1 CH933810 KRF94150.1 GEDC01001166 JAS36132.1 GEDC01008789 JAS28509.1 ODYU01006758 SOQ48894.1 KQ978457 KYM93875.1 JTDY01001535 KOB73623.1 CM000070 EAL28208.2 KB632093 ERL88690.1 APGK01017806 APGK01017807 APGK01017808 KB740053 ENN81797.1 CH479179 EDW25146.1 GBXI01014482 GBXI01005217 JAC99809.1 JAD09075.1 GBXI01008342 JAD05950.1 AXCM01003686 KQ983136 KYQ47144.1 GAKP01004275 JAC54677.1 LJIJ01000233 ODN00115.1 GFPF01008342 MAA19488.1 ADMH02001351 ETN62915.1 RSAL01000038 RVE51080.1 KRF94153.1 KRF94156.1 KRF94154.1 ATLV01016989 ATLV01016990 KE525139 KFB41706.1 GEDV01008732 JAP79825.1 KRF94152.1 AJVK01007778 AJVK01007779 AJVK01007780 QOIP01000008 RLU19726.1 GAMC01020746 JAB85809.1 KRF94157.1 KRF94149.1 KRF94155.1 KRF94151.1 GAMC01020747 JAB85808.1 GAMC01020748 JAB85807.1 GEDV01000917 JAP87640.1 KQ425015 KOF70121.1 ABLF02033685 ABLF02033689 ABLF02033690 HACG01037197 CEK84062.1 GGFK01006333 MBW39654.1 GGFK01006544 MBW39865.1 EDW07699.2 GAKP01004273 JAC54679.1 GEZM01023937 GEZM01023935 GEZM01023934 JAV88128.1 GEZM01023936 GEZM01023933 GEZM01023932 JAV88130.1 KQ422575 KOF74633.1 KOF74632.1 GGFK01000644 MBW33965.1 ADTU01021233 ADTU01021234 ADTU01021235 ADTU01021236 ADTU01021237 ADTU01021238 ADTU01021239 ADTU01021240 ADTU01021241 ADTU01021242 GU454802 ADK55596.1 GBXI01009541 JAD04751.1 ATLV01017677 ATLV01017678 ATLV01017679 KE525181 KFB42386.1 KQ981208 KYN44709.1 EU024890 ABW79798.1 GEFH01001080 JAP67501.1 JI181595 ADY48637.1 JI167980 ADY42882.1 GGFJ01000077 MBW49218.1 AXCN02001967 JI168330 ADY43125.1 GDHF01030583 GDHF01015320 JAI21731.1 JAI36994.1 KZ288195 PBC33792.1 KQ420694 KOF79592.1 KQ765323 KQ759877 OAD53988.1 OAD62273.1 GFPF01008343 MAA19489.1 GBBK01005163 JAC19319.1 GEDV01008731 JAP79826.1 GFPF01008344 MAA19490.1 GEDV01000919 JAP87638.1 GDHF01011599 JAI40715.1 GEDV01000918 JAP87639.1 AHAT01019149 AHAT01019150 AHAT01019151 NEDP02005415 OWF41229.1 KQ435760 KOX75591.1 KQ460953 KPJ10291.1 KQ773953 OAD52336.1 KQ459605 KPI92029.1 AXCN02000208 GAMD01001822 JAA99768.1 JR043982 AEY59824.1 AMQN01008945 KB304240 ELU02214.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000218220
UP000078492
+ More
UP000009192 UP000078542 UP000037510 UP000001819 UP000030742 UP000019118 UP000008744 UP000075883 UP000075809 UP000094527 UP000000673 UP000283053 UP000030765 UP000092462 UP000279307 UP000053454 UP000007819 UP000265140 UP000069272 UP000005205 UP000261600 UP000078541 UP000075886 UP000242457 UP000079169 UP000018468 UP000242188 UP000053105 UP000075880 UP000014760
UP000009192 UP000078542 UP000037510 UP000001819 UP000030742 UP000019118 UP000008744 UP000075883 UP000075809 UP000094527 UP000000673 UP000283053 UP000030765 UP000092462 UP000279307 UP000053454 UP000007819 UP000265140 UP000069272 UP000005205 UP000261600 UP000078541 UP000075886 UP000242457 UP000079169 UP000018468 UP000242188 UP000053105 UP000075880 UP000014760
Pfam
PF00084 Sushi
+ More
PF00057 Ldl_recept_a
PF00089 Trypsin
PF07686 V-set
PF00052 Laminin_B
PF00008 EGF
PF00054 Laminin_G_1
PF00053 Laminin_EGF
PF13895 Ig_2
PF02210 Laminin_G_2
PF07679 I-set
PF00047 ig
PF01392 Fz
PF00041 fn3
PF15902 Sortilin-Vps10
PF00058 Ldl_recept_b
PF15901 Sortilin_C
PF01390 SEA
PF07645 EGF_CA
PF12662 cEGF
PF00004 AAA
PF01416 PseudoU_synth_1
PF00439 Bromodomain
PF00431 CUB
PF00057 Ldl_recept_a
PF00089 Trypsin
PF07686 V-set
PF00052 Laminin_B
PF00008 EGF
PF00054 Laminin_G_1
PF00053 Laminin_EGF
PF13895 Ig_2
PF02210 Laminin_G_2
PF07679 I-set
PF00047 ig
PF01392 Fz
PF00041 fn3
PF15902 Sortilin-Vps10
PF00058 Ldl_recept_b
PF15901 Sortilin_C
PF01390 SEA
PF07645 EGF_CA
PF12662 cEGF
PF00004 AAA
PF01416 PseudoU_synth_1
PF00439 Bromodomain
PF00431 CUB
Interpro
IPR023415
LDLR_class-A_CS
+ More
IPR000436 Sushi_SCR_CCP_dom
IPR018114 TRYPSIN_HIS
IPR001254 Trypsin_dom
IPR009003 Peptidase_S1_PA
IPR002172 LDrepeatLR_classA_rpt
IPR036055 LDL_receptor-like_sf
IPR035976 Sushi/SCR/CCP_sf
IPR013106 Ig_V-set
IPR000742 EGF-like_dom
IPR001791 Laminin_G
IPR036179 Ig-like_dom_sf
IPR003598 Ig_sub2
IPR013032 EGF-like_CS
IPR013783 Ig-like_fold
IPR000034 Laminin_IV
IPR002049 Laminin_EGF
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR013098 Ig_I-set
IPR013320 ConA-like_dom_sf
IPR001881 EGF-like_Ca-bd_dom
IPR000082 SEA_dom
IPR013151 Immunoglobulin
IPR040093 LRAD1
IPR036790 Frizzled_dom_sf
IPR020067 Frizzled_dom
IPR009030 Growth_fac_rcpt_cys_sf
IPR011042 6-blade_b-propeller_TolB-like
IPR006581 VPS10
IPR031777 Sortilin_C
IPR031778 Sortilin_N
IPR036116 FN3_sf
IPR000033 LDLR_classB_rpt
IPR003961 FN3_dom
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR036364 SEA_dom_sf
IPR018097 EGF_Ca-bd_CS
IPR026823 cEGF
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR032931 VLDLR
IPR003593 AAA+_ATPase
IPR001487 Bromodomain
IPR020095 PsdUridine_synth_TruA_C
IPR003959 ATPase_AAA_core
IPR020103 PsdUridine_synth_cat_dom_sf
IPR036427 Bromodomain-like_sf
IPR001406 PsdUridine_synth_TruA
IPR020097 PsdUridine_synth_TruA_a/b_dom
IPR041708 PUS1/PUS2-like
IPR027417 P-loop_NTPase
IPR003960 ATPase_AAA_CS
IPR035914 Sperma_CUB_dom_sf
IPR000859 CUB_dom
IPR033116 TRYPSIN_SER
IPR000436 Sushi_SCR_CCP_dom
IPR018114 TRYPSIN_HIS
IPR001254 Trypsin_dom
IPR009003 Peptidase_S1_PA
IPR002172 LDrepeatLR_classA_rpt
IPR036055 LDL_receptor-like_sf
IPR035976 Sushi/SCR/CCP_sf
IPR013106 Ig_V-set
IPR000742 EGF-like_dom
IPR001791 Laminin_G
IPR036179 Ig-like_dom_sf
IPR003598 Ig_sub2
IPR013032 EGF-like_CS
IPR013783 Ig-like_fold
IPR000034 Laminin_IV
IPR002049 Laminin_EGF
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR013098 Ig_I-set
IPR013320 ConA-like_dom_sf
IPR001881 EGF-like_Ca-bd_dom
IPR000082 SEA_dom
IPR013151 Immunoglobulin
IPR040093 LRAD1
IPR036790 Frizzled_dom_sf
IPR020067 Frizzled_dom
IPR009030 Growth_fac_rcpt_cys_sf
IPR011042 6-blade_b-propeller_TolB-like
IPR006581 VPS10
IPR031777 Sortilin_C
IPR031778 Sortilin_N
IPR036116 FN3_sf
IPR000033 LDLR_classB_rpt
IPR003961 FN3_dom
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR036364 SEA_dom_sf
IPR018097 EGF_Ca-bd_CS
IPR026823 cEGF
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR032931 VLDLR
IPR003593 AAA+_ATPase
IPR001487 Bromodomain
IPR020095 PsdUridine_synth_TruA_C
IPR003959 ATPase_AAA_core
IPR020103 PsdUridine_synth_cat_dom_sf
IPR036427 Bromodomain-like_sf
IPR001406 PsdUridine_synth_TruA
IPR020097 PsdUridine_synth_TruA_a/b_dom
IPR041708 PUS1/PUS2-like
IPR027417 P-loop_NTPase
IPR003960 ATPase_AAA_CS
IPR035914 Sperma_CUB_dom_sf
IPR000859 CUB_dom
IPR033116 TRYPSIN_SER
SUPFAM
Gene 3D
CDD
ProteinModelPortal
I3WAD0
H9JRQ5
A0A0N0PDF1
A0A2H1VKP2
Q69BL0
A0A194Q0X1
+ More
A0A212FFN5 A0A2W1BIK5 A0A2W1BIE4 A0A2A4J8A8 A0A1B6CKY8 A0A195E4R2 A0A0Q9WWR1 A0A1B6EDX6 A0A1B6DS91 A0A2H1W8T0 A0A195BZV6 A0A0L7LDM0 Q297J7 U4UFJ4 N6USS0 B4G4A8 A0A0A1XDP6 A0A0A1X5C7 A0A182LSY4 A0A151WH43 A0A034WK22 A0A1D2N4X8 A0A224YPF5 W5JJ60 A0A3S2M4C7 A0A0Q9WYR9 A0A0Q9WV89 A0A0Q9X050 A0A084VUR2 A0A131YML4 A0A0Q9WN73 A0A1B0F0M3 A0A3L8DHC7 W8B7J5 A0A0Q9WN66 A0A0Q9X076 A0A0Q9WPG5 A0A0Q9WNJ7 W8AMV9 W8AJ92 A0A131Z9Z9 A0A0L8FZG5 J9K8R8 A0A0B7ATX1 A0A2M4AFU1 A0A2M4AGF6 B4L1X8 A0A3P9A5H3 A0A034WJ53 A0A1Y1MSQ2 A0A1Y1MUH0 A0A182F8Q3 A0A0L8GCJ0 A0A0L8GC81 A0A2M3ZZN1 A0A158NND7 G8Z9Y7 A0A0A1X1K7 A0A3Q3KPP9 A0A084VWP2 A0A195FXZ6 A8TEF0 A0A131XPK7 F1LET3 F1KYC8 A0A2M4B8R2 A0A182QRH4 F1KZ21 A0A0K8VDH4 A0A2A3ER96 A0A0L8GRS2 A0A3Q0JGI2 A0A310SF19 A0A224YUS3 A0A023FEK7 A0A131YP00 A0A224Z0F1 A0A131Z9J2 A0A0K8VQ98 A0A131Z8F7 W5M8L7 A0A210PXP2 A0A0N0BH65 A0A194R369 A0A310SBQ4 A0A182ITD4 A0A194PH43 A0A182Q107 T1E8X0 V9IHL7 R7U896
A0A212FFN5 A0A2W1BIK5 A0A2W1BIE4 A0A2A4J8A8 A0A1B6CKY8 A0A195E4R2 A0A0Q9WWR1 A0A1B6EDX6 A0A1B6DS91 A0A2H1W8T0 A0A195BZV6 A0A0L7LDM0 Q297J7 U4UFJ4 N6USS0 B4G4A8 A0A0A1XDP6 A0A0A1X5C7 A0A182LSY4 A0A151WH43 A0A034WK22 A0A1D2N4X8 A0A224YPF5 W5JJ60 A0A3S2M4C7 A0A0Q9WYR9 A0A0Q9WV89 A0A0Q9X050 A0A084VUR2 A0A131YML4 A0A0Q9WN73 A0A1B0F0M3 A0A3L8DHC7 W8B7J5 A0A0Q9WN66 A0A0Q9X076 A0A0Q9WPG5 A0A0Q9WNJ7 W8AMV9 W8AJ92 A0A131Z9Z9 A0A0L8FZG5 J9K8R8 A0A0B7ATX1 A0A2M4AFU1 A0A2M4AGF6 B4L1X8 A0A3P9A5H3 A0A034WJ53 A0A1Y1MSQ2 A0A1Y1MUH0 A0A182F8Q3 A0A0L8GCJ0 A0A0L8GC81 A0A2M3ZZN1 A0A158NND7 G8Z9Y7 A0A0A1X1K7 A0A3Q3KPP9 A0A084VWP2 A0A195FXZ6 A8TEF0 A0A131XPK7 F1LET3 F1KYC8 A0A2M4B8R2 A0A182QRH4 F1KZ21 A0A0K8VDH4 A0A2A3ER96 A0A0L8GRS2 A0A3Q0JGI2 A0A310SF19 A0A224YUS3 A0A023FEK7 A0A131YP00 A0A224Z0F1 A0A131Z9J2 A0A0K8VQ98 A0A131Z8F7 W5M8L7 A0A210PXP2 A0A0N0BH65 A0A194R369 A0A310SBQ4 A0A182ITD4 A0A194PH43 A0A182Q107 T1E8X0 V9IHL7 R7U896
PDB
3M0C
E-value=3.63938e-07,
Score=123
Ontologies
KEGG
GO
GO:0004252
GO:0016021
GO:0005509
GO:0045752
GO:0050832
GO:0050830
GO:0045087
GO:0005794
GO:0005604
GO:2001262
GO:0008045
GO:0007415
GO:0040037
GO:0045199
GO:0072347
GO:0005524
GO:0003723
GO:0009982
GO:0001522
GO:0005515
GO:0008198
GO:0016491
GO:0008270
GO:0016701
GO:0006355
GO:0003824
GO:0008152
GO:0015930
PANTHER
Topology
Subcellular location
Golgi apparatus
SignalP
Position: 1 - 17,
Likelihood: 0.997283
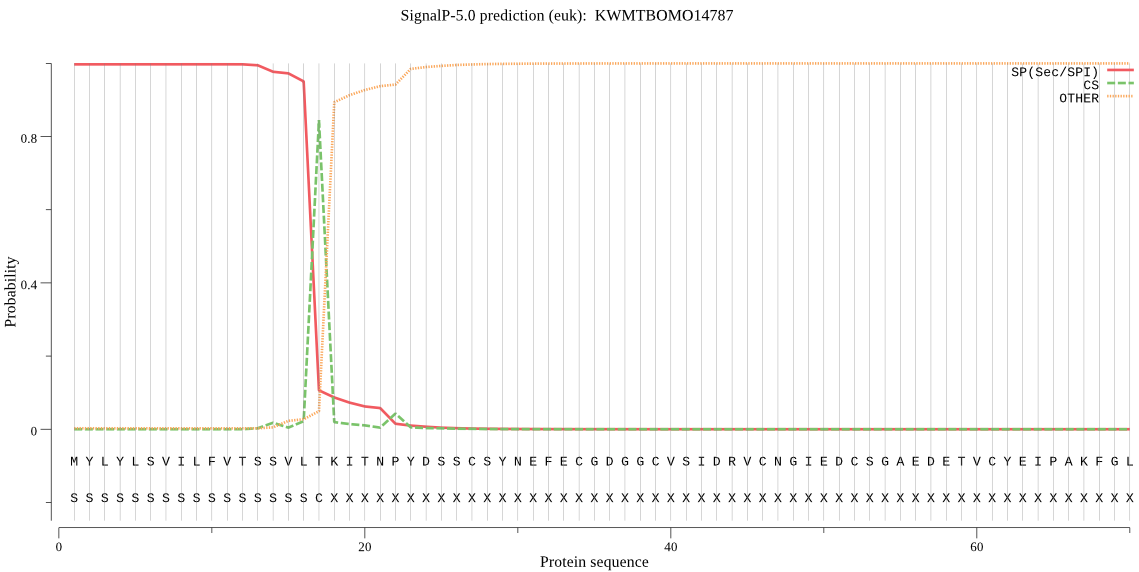
Length:
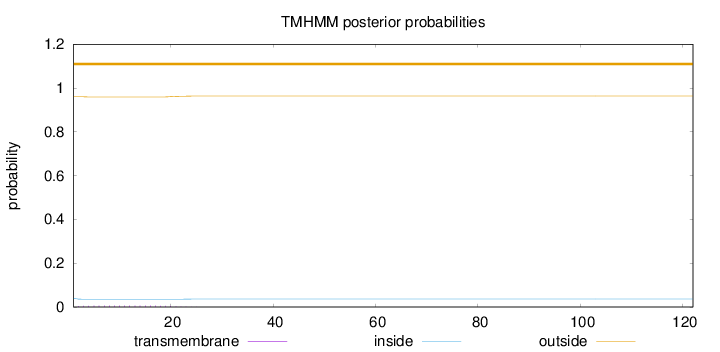
122
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.11562
Exp number, first 60 AAs:
0.11562
Total prob of N-in:
0.04021
outside
1 - 122
Population Genetic Test Statistics
Pi
194.247832
Theta
164.554278
Tajima's D
0.037554
CLR
1126.311168
CSRT
0.375981200939953
Interpretation
Uncertain
