Gene
KWMTBOMO14777
Pre Gene Modal
BGIBMGA003841
Annotation
PREDICTED:_transferrin-like_[Bombyx_mori]
Full name
Transferrin
Location in the cell
Extracellular Reliability : 1.351
Sequence
CDS
ATGCCAGTTTTTAGTAGTGTGTTAGTTTTTTTTTCTTTATTAAACGTTGTCTTGCTACAAAGAACAGGATCCGGAACGAATGGGAACTTGCGTCTGTGTGTTGTGGAAGGCCGCGGCTCGTACAAACGGGCCCCCAAGTTCTGTCCCGTGCTGGACGCCGAGGATTCGGGGGCGGAGTGCGTCATTGGAACCGACAGGCTGGACTGTCTCCGCCGCATCCACAAGGGGACCGTTGACTTCGGCGTCTTCAGCCCCGAGGATCTGGTCGCAGCGCAATGGGCCAACGTTGACGTTCTGGTCACCAACGAACTCCGGATGAGAGCCAGACCCTTCGAACGCAGCGTCGTTGCGGTAGTGAACAGAAAGATCCTGCCGGATAGTTCCACTTCGGTCCACGCGGTGCTCCGGAATACAACGCTGTGCCACCCGGGAGTCGGGGTCGATGACATCCGGCCCCTGAGCGACACACTGTCCGGGTATTTAGAATCTCTAGTATTGGAAAGATCCTGCGACCCGGATCTGTCGCTGACCGAGAACAGAGTGAAGTCGTTGTCCGAATACTTTGGGAAGGCCTGCAAGGCGGGGGCGTGGCTACCGGACAGCACCAGAGACGCTGAACTTAAGCGTAAATATTCGTCAATGTGCGCGGCCTGCGGTAACCCATCCAAGTGTAGCGCTTCGGACGTGTACTGGGGTAACAGCGGCGCTCTCGCGTGCCTCGGTAACGGAGCCGGTGACGTCACTTGGGCGGAACTGGACGATGTTAACTCGTATTTTGCGCTTACACCATCGACAAACCCACCCAGTTCTGCAGCTGATCAGTTCGCGTATCTCTGCCGAGACGGTACCTGGCAGGACCTCAATAACAACCCTGAGCCTTGCGTCTGGCTGCACAGACCTTGGCCAGTAATTGTCGCTAAAAAGAAGGCCGCAGAAGCAGTAGCAAGCTTAGACAGAATCTTAACGAACACCAGCATCACGGTGGACCAGCATTGGCGCGGGGCCCTGGCCGCTCTGATGGAGATCCACCAAGCTCAGCCCGAGAAGCTTCAACCTCCTAAAGCTCCTTTGGATTATTTGGCCAGAGCCAAGGGCTTTAGAGAGGCGTATAGTCAAACCGGATGTGATCCTCCCAGGTTCATTACTCTATGCACTACATCATTTCTGGCGAAGGCGAAATGCGAGTGGCTTAGCGAAGCCGGAGCTGTCTACGGTATAGCCCCACCTCTTCAGTGCGTAGTTAGAAAAGACATGCAAGAGTGTATGAAGTCGGTCAGCATTGGGGAGTCTGACGTCACAGTTGGGAACAGTGACTGGCTTGTCAAAGGGAAAAGAGATTTGTCTATAGCGCCGGTGCTGCACGAAGCGACACCTATCGTCGAGAAAACGAGCACAGTTGTGGCTTACGTCAGAAAGGACTCAGCAATAAGCAGTATGGTGGACTTGCGCGGTAAGAAGGCGGCGTTCCCTCGCCACGATGGCGCCGCCTGGCATTCCGTACTGCGCTACATTACTGAAAATCAAAAGATCACCTGCAGCGATCTCGTCGGAGAGTATTTCGAGGAGATCTGCGCTCCGGGTGTCGGGGATCAGAATCCCGGTGGCAAATTAAAGGATTGCTACGAAGAAGGGGAGATGGATCCCCTCCGCTACCTCGCTGAAGGCAAAACTGATGTTGCTTTCGTCAGTATTAATAGTTTTAATTTGTTCAAAGGAATGCCAAGTATGGACGCTTCCAACATCAGACCGCTGTGTCCGGACGAGAACCAAATGTACTGCTTCATAAGTTGGTCCAATGTTGGCCACGTGTTCGCCGCCAATAACGTAACTGACAGCCGAAGGCACGAGATCGTAAACGTAATGACGAAACTGGACCAACTGTTCGGCAAACATCCCCCATTCCACAACCCGATGTTCTCAATGTACGGCGCCTTCAATCACGAGATGGACGTTATATTCCACAATAACACGAAAACGTTGGCCACGGACGCCATGTTGGCAATGCATCCGTATGATAAAATACCAATGAATTTTGAGCGGGAGATCTCGAAGATGACCGATTTTGTTTGCGGTTACGGCGTCAATATGACACCTTCTTTGTTTTTATTAATAAGTCTAGTCGTTTTTGTGATTTTAAGATAA
Protein
MPVFSSVLVFFSLLNVVLLQRTGSGTNGNLRLCVVEGRGSYKRAPKFCPVLDAEDSGAECVIGTDRLDCLRRIHKGTVDFGVFSPEDLVAAQWANVDVLVTNELRMRARPFERSVVAVVNRKILPDSSTSVHAVLRNTTLCHPGVGVDDIRPLSDTLSGYLESLVLERSCDPDLSLTENRVKSLSEYFGKACKAGAWLPDSTRDAELKRKYSSMCAACGNPSKCSASDVYWGNSGALACLGNGAGDVTWAELDDVNSYFALTPSTNPPSSAADQFAYLCRDGTWQDLNNNPEPCVWLHRPWPVIVAKKKAAEAVASLDRILTNTSITVDQHWRGALAALMEIHQAQPEKLQPPKAPLDYLARAKGFREAYSQTGCDPPRFITLCTTSFLAKAKCEWLSEAGAVYGIAPPLQCVVRKDMQECMKSVSIGESDVTVGNSDWLVKGKRDLSIAPVLHEATPIVEKTSTVVAYVRKDSAISSMVDLRGKKAAFPRHDGAAWHSVLRYITENQKITCSDLVGEYFEEICAPGVGDQNPGGKLKDCYEEGEMDPLRYLAEGKTDVAFVSINSFNLFKGMPSMDASNIRPLCPDENQMYCFISWSNVGHVFAANNVTDSRRHEIVNVMTKLDQLFGKHPPFHNPMFSMYGAFNHEMDVIFHNNTKTLATDAMLAMHPYDKIPMNFEREISKMTDFVCGYGVNMTPSLFLLISLVVFVILR
Summary
Description
Transferrins are iron binding transport proteins which bind Fe(3+) ion in association with the binding of an anion, usually bicarbonate.
Similarity
Belongs to the transferrin family.
Feature
chain Transferrin
Uniprot
H9J2V4
A0A2H1VG88
A0A2A4J6R4
A0A3S2NUK7
A0A194Q0Z9
A0A212EZE6
+ More
B3NPU9 B4P6B5 A0A0Q9X9H9 A0A1I8NYX9 B4KPS1 A1ZAC0 A0A0M4EHV7 A0A0J9RDR5 A0A1W4VE03 A0A0Q9X9I0 B4HST7 A0A3B0JCE1 B4MKD8 A0A1I8M8Y0 B3MH63 B4J5J2 B4GI86 Q28Z61 B4LKY0 A0A1Y1NDV8 A0A1B0BXC0 A0A182JS15 A0A1A9ZBI4 A0A182QQU3 A0A0A1WYR2 A0A075BIF7 A0A0K8VZ27 A0A034WCT0 A0A1B0GBJ1 A0A1A9W0T6 W8BBR2 A0A0Q9W3H8 A0A182J5J5 A0A182N2C7 A0A0L0CN40 A0A182HWD4 Q16KL8 A0A1S4FXI3 A0A182VH29 A0A084VHV0 A0A182YB33 A0A182X5L5 A0A182NZL2 Q7QHF6 A0A1W7R6N2 A0A182UCH4 A0A1W4X7C9 A0A182GHI4 A0A182RTE1 A0A182WDH7 A0A182FKJ6 A0A182MWS7 A0A1B0D8P7 A0A1A9VE39 W5J913 A0A1V1FNP4 A0A067RHB7 A0A2J7RC93 A0A139WNS1 T1GUA6 A0A0T6BCJ2 A0A1A9XED1 A0A182T5K6 A0A2J7RC83 A0A336LVW1 U4UIH4 A0A158NUB5 A0A2S2QHV0 A0A2J7RC86 A0A2S2PJB6 J9JLU1 A0A088A4K4 A0A2H8TQM1 A0A195FKE4 A0A151IXL4 A0A026WEV5 A0A195BBQ3 A0A151ILR0 A0A151WEY5 A0A2P8YP82 F4WR43 A0A2A3EET7 E2APS4 A0A2Z5BRU1 A0A0L7QMM2 A0A0C9R5K1
B3NPU9 B4P6B5 A0A0Q9X9H9 A0A1I8NYX9 B4KPS1 A1ZAC0 A0A0M4EHV7 A0A0J9RDR5 A0A1W4VE03 A0A0Q9X9I0 B4HST7 A0A3B0JCE1 B4MKD8 A0A1I8M8Y0 B3MH63 B4J5J2 B4GI86 Q28Z61 B4LKY0 A0A1Y1NDV8 A0A1B0BXC0 A0A182JS15 A0A1A9ZBI4 A0A182QQU3 A0A0A1WYR2 A0A075BIF7 A0A0K8VZ27 A0A034WCT0 A0A1B0GBJ1 A0A1A9W0T6 W8BBR2 A0A0Q9W3H8 A0A182J5J5 A0A182N2C7 A0A0L0CN40 A0A182HWD4 Q16KL8 A0A1S4FXI3 A0A182VH29 A0A084VHV0 A0A182YB33 A0A182X5L5 A0A182NZL2 Q7QHF6 A0A1W7R6N2 A0A182UCH4 A0A1W4X7C9 A0A182GHI4 A0A182RTE1 A0A182WDH7 A0A182FKJ6 A0A182MWS7 A0A1B0D8P7 A0A1A9VE39 W5J913 A0A1V1FNP4 A0A067RHB7 A0A2J7RC93 A0A139WNS1 T1GUA6 A0A0T6BCJ2 A0A1A9XED1 A0A182T5K6 A0A2J7RC83 A0A336LVW1 U4UIH4 A0A158NUB5 A0A2S2QHV0 A0A2J7RC86 A0A2S2PJB6 J9JLU1 A0A088A4K4 A0A2H8TQM1 A0A195FKE4 A0A151IXL4 A0A026WEV5 A0A195BBQ3 A0A151ILR0 A0A151WEY5 A0A2P8YP82 F4WR43 A0A2A3EET7 E2APS4 A0A2Z5BRU1 A0A0L7QMM2 A0A0C9R5K1
Pubmed
19121390
26354079
22118469
17994087
17550304
10731132
+ More
12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25315136 15632085 28004739 25830018 25348373 24495485 26108605 17510324 24438588 25244985 12364791 26483478 20920257 23761445 28410430 24845553 18362917 19820115 23537049 21347285 24508170 30249741 29403074 21719571 20798317 29272461
12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25315136 15632085 28004739 25830018 25348373 24495485 26108605 17510324 24438588 25244985 12364791 26483478 20920257 23761445 28410430 24845553 18362917 19820115 23537049 21347285 24508170 30249741 29403074 21719571 20798317 29272461
EMBL
BABH01036991
BABH01036992
BABH01036993
BABH01036994
ODYU01002408
SOQ39845.1
+ More
NWSH01002862 PCG67426.1 RSAL01000072 RVE49064.1 KQ459580 KPI99226.1 AGBW02011291 OWR46876.1 CH954179 EDV55796.1 CM000158 EDW91965.1 CH933808 KRG05088.1 EDW10198.1 AE013599 BT099700 AAF58039.1 ACV53064.1 CP012524 ALC42235.1 CM002911 KMY94213.1 KRG05087.1 CH480816 EDW48101.1 OUUW01000001 SPP72890.1 CH963846 EDW72577.1 CH902619 EDV35822.1 CH916367 EDW00755.1 CH479183 EDW36206.1 CM000071 EAL25753.2 CH940648 EDW60784.1 GEZM01005633 JAV96021.1 JXJN01022162 AXCN02001728 GBXI01010088 JAD04204.1 JX418003 AGL91668.1 GDHF01008146 JAI44168.1 GAKP01006463 JAC52489.1 CCAG010014016 GAMC01015984 JAB90571.1 KRF79619.1 JRES01000264 KNC32859.1 APCN01001485 CH477951 EAT34844.1 ATLV01013235 KE524847 KFB37544.1 AAAB01008816 EAA05142.3 GEHC01000852 JAV46793.1 JXUM01011789 JXUM01011790 KQ560338 KXJ82966.1 AXCM01000599 AJVK01027629 AJVK01027630 ADMH02002090 ETN59370.1 FX985457 BAX07470.1 KK852636 KDR19727.1 NEVH01005885 PNF38438.1 KQ971311 KYB29461.1 CAQQ02190360 LJIG01001874 KRT85066.1 PNF38439.1 UFQT01000177 SSX21181.1 KB632314 ERL92178.1 ADTU01026277 ADTU01026278 ADTU01026279 GGMS01008115 MBY77318.1 PNF38440.1 GGMR01016914 MBY29533.1 ABLF02022105 ABLF02022109 ABLF02022110 GFXV01004117 MBW15922.1 KQ981512 KYN40873.1 KQ980807 KYN12736.1 KK107238 QOIP01000007 EZA54592.1 RLU20819.1 KQ976532 KYM81652.1 KQ977104 KYN05803.1 KQ983238 KYQ46345.1 PYGN01000456 PSN46014.1 GL888284 EGI63270.1 KZ288266 PBC30247.1 GL441581 EFN64550.1 MF627746 AXA98486.1 KQ414885 KOC59867.1 GBYB01002101 JAG71868.1
NWSH01002862 PCG67426.1 RSAL01000072 RVE49064.1 KQ459580 KPI99226.1 AGBW02011291 OWR46876.1 CH954179 EDV55796.1 CM000158 EDW91965.1 CH933808 KRG05088.1 EDW10198.1 AE013599 BT099700 AAF58039.1 ACV53064.1 CP012524 ALC42235.1 CM002911 KMY94213.1 KRG05087.1 CH480816 EDW48101.1 OUUW01000001 SPP72890.1 CH963846 EDW72577.1 CH902619 EDV35822.1 CH916367 EDW00755.1 CH479183 EDW36206.1 CM000071 EAL25753.2 CH940648 EDW60784.1 GEZM01005633 JAV96021.1 JXJN01022162 AXCN02001728 GBXI01010088 JAD04204.1 JX418003 AGL91668.1 GDHF01008146 JAI44168.1 GAKP01006463 JAC52489.1 CCAG010014016 GAMC01015984 JAB90571.1 KRF79619.1 JRES01000264 KNC32859.1 APCN01001485 CH477951 EAT34844.1 ATLV01013235 KE524847 KFB37544.1 AAAB01008816 EAA05142.3 GEHC01000852 JAV46793.1 JXUM01011789 JXUM01011790 KQ560338 KXJ82966.1 AXCM01000599 AJVK01027629 AJVK01027630 ADMH02002090 ETN59370.1 FX985457 BAX07470.1 KK852636 KDR19727.1 NEVH01005885 PNF38438.1 KQ971311 KYB29461.1 CAQQ02190360 LJIG01001874 KRT85066.1 PNF38439.1 UFQT01000177 SSX21181.1 KB632314 ERL92178.1 ADTU01026277 ADTU01026278 ADTU01026279 GGMS01008115 MBY77318.1 PNF38440.1 GGMR01016914 MBY29533.1 ABLF02022105 ABLF02022109 ABLF02022110 GFXV01004117 MBW15922.1 KQ981512 KYN40873.1 KQ980807 KYN12736.1 KK107238 QOIP01000007 EZA54592.1 RLU20819.1 KQ976532 KYM81652.1 KQ977104 KYN05803.1 KQ983238 KYQ46345.1 PYGN01000456 PSN46014.1 GL888284 EGI63270.1 KZ288266 PBC30247.1 GL441581 EFN64550.1 MF627746 AXA98486.1 KQ414885 KOC59867.1 GBYB01002101 JAG71868.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000007151
UP000008711
+ More
UP000002282 UP000009192 UP000095300 UP000000803 UP000092553 UP000192221 UP000001292 UP000268350 UP000007798 UP000095301 UP000007801 UP000001070 UP000008744 UP000001819 UP000008792 UP000092460 UP000075881 UP000092445 UP000075886 UP000092444 UP000091820 UP000075880 UP000075884 UP000037069 UP000075840 UP000008820 UP000075903 UP000030765 UP000076408 UP000076407 UP000075885 UP000007062 UP000075902 UP000192223 UP000069940 UP000249989 UP000075900 UP000075920 UP000069272 UP000075883 UP000092462 UP000078200 UP000000673 UP000027135 UP000235965 UP000007266 UP000015102 UP000092443 UP000075901 UP000030742 UP000005205 UP000007819 UP000005203 UP000078541 UP000078492 UP000053097 UP000279307 UP000078540 UP000078542 UP000075809 UP000245037 UP000007755 UP000242457 UP000000311 UP000053825
UP000002282 UP000009192 UP000095300 UP000000803 UP000092553 UP000192221 UP000001292 UP000268350 UP000007798 UP000095301 UP000007801 UP000001070 UP000008744 UP000001819 UP000008792 UP000092460 UP000075881 UP000092445 UP000075886 UP000092444 UP000091820 UP000075880 UP000075884 UP000037069 UP000075840 UP000008820 UP000075903 UP000030765 UP000076408 UP000076407 UP000075885 UP000007062 UP000075902 UP000192223 UP000069940 UP000249989 UP000075900 UP000075920 UP000069272 UP000075883 UP000092462 UP000078200 UP000000673 UP000027135 UP000235965 UP000007266 UP000015102 UP000092443 UP000075901 UP000030742 UP000005205 UP000007819 UP000005203 UP000078541 UP000078492 UP000053097 UP000279307 UP000078540 UP000078542 UP000075809 UP000245037 UP000007755 UP000242457 UP000000311 UP000053825
Interpro
IPR002110
Ankyrin_rpt
+ More
IPR001156 Transferrin-like_dom
IPR001357 BRCT_dom
IPR036420 BRCT_dom_sf
IPR036770 Ankyrin_rpt-contain_sf
IPR020683 Ankyrin_rpt-contain_dom
IPR016357 Transferrin
IPR032675 LRR_dom_sf
IPR000182 GNAT_dom
IPR016181 Acyl_CoA_acyltransferase
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR001680 WD40_repeat
IPR036322 WD40_repeat_dom_sf
IPR019775 WD40_repeat_CS
IPR001156 Transferrin-like_dom
IPR001357 BRCT_dom
IPR036420 BRCT_dom_sf
IPR036770 Ankyrin_rpt-contain_sf
IPR020683 Ankyrin_rpt-contain_dom
IPR016357 Transferrin
IPR032675 LRR_dom_sf
IPR000182 GNAT_dom
IPR016181 Acyl_CoA_acyltransferase
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR001680 WD40_repeat
IPR036322 WD40_repeat_dom_sf
IPR019775 WD40_repeat_CS
Gene 3D
ProteinModelPortal
H9J2V4
A0A2H1VG88
A0A2A4J6R4
A0A3S2NUK7
A0A194Q0Z9
A0A212EZE6
+ More
B3NPU9 B4P6B5 A0A0Q9X9H9 A0A1I8NYX9 B4KPS1 A1ZAC0 A0A0M4EHV7 A0A0J9RDR5 A0A1W4VE03 A0A0Q9X9I0 B4HST7 A0A3B0JCE1 B4MKD8 A0A1I8M8Y0 B3MH63 B4J5J2 B4GI86 Q28Z61 B4LKY0 A0A1Y1NDV8 A0A1B0BXC0 A0A182JS15 A0A1A9ZBI4 A0A182QQU3 A0A0A1WYR2 A0A075BIF7 A0A0K8VZ27 A0A034WCT0 A0A1B0GBJ1 A0A1A9W0T6 W8BBR2 A0A0Q9W3H8 A0A182J5J5 A0A182N2C7 A0A0L0CN40 A0A182HWD4 Q16KL8 A0A1S4FXI3 A0A182VH29 A0A084VHV0 A0A182YB33 A0A182X5L5 A0A182NZL2 Q7QHF6 A0A1W7R6N2 A0A182UCH4 A0A1W4X7C9 A0A182GHI4 A0A182RTE1 A0A182WDH7 A0A182FKJ6 A0A182MWS7 A0A1B0D8P7 A0A1A9VE39 W5J913 A0A1V1FNP4 A0A067RHB7 A0A2J7RC93 A0A139WNS1 T1GUA6 A0A0T6BCJ2 A0A1A9XED1 A0A182T5K6 A0A2J7RC83 A0A336LVW1 U4UIH4 A0A158NUB5 A0A2S2QHV0 A0A2J7RC86 A0A2S2PJB6 J9JLU1 A0A088A4K4 A0A2H8TQM1 A0A195FKE4 A0A151IXL4 A0A026WEV5 A0A195BBQ3 A0A151ILR0 A0A151WEY5 A0A2P8YP82 F4WR43 A0A2A3EET7 E2APS4 A0A2Z5BRU1 A0A0L7QMM2 A0A0C9R5K1
B3NPU9 B4P6B5 A0A0Q9X9H9 A0A1I8NYX9 B4KPS1 A1ZAC0 A0A0M4EHV7 A0A0J9RDR5 A0A1W4VE03 A0A0Q9X9I0 B4HST7 A0A3B0JCE1 B4MKD8 A0A1I8M8Y0 B3MH63 B4J5J2 B4GI86 Q28Z61 B4LKY0 A0A1Y1NDV8 A0A1B0BXC0 A0A182JS15 A0A1A9ZBI4 A0A182QQU3 A0A0A1WYR2 A0A075BIF7 A0A0K8VZ27 A0A034WCT0 A0A1B0GBJ1 A0A1A9W0T6 W8BBR2 A0A0Q9W3H8 A0A182J5J5 A0A182N2C7 A0A0L0CN40 A0A182HWD4 Q16KL8 A0A1S4FXI3 A0A182VH29 A0A084VHV0 A0A182YB33 A0A182X5L5 A0A182NZL2 Q7QHF6 A0A1W7R6N2 A0A182UCH4 A0A1W4X7C9 A0A182GHI4 A0A182RTE1 A0A182WDH7 A0A182FKJ6 A0A182MWS7 A0A1B0D8P7 A0A1A9VE39 W5J913 A0A1V1FNP4 A0A067RHB7 A0A2J7RC93 A0A139WNS1 T1GUA6 A0A0T6BCJ2 A0A1A9XED1 A0A182T5K6 A0A2J7RC83 A0A336LVW1 U4UIH4 A0A158NUB5 A0A2S2QHV0 A0A2J7RC86 A0A2S2PJB6 J9JLU1 A0A088A4K4 A0A2H8TQM1 A0A195FKE4 A0A151IXL4 A0A026WEV5 A0A195BBQ3 A0A151ILR0 A0A151WEY5 A0A2P8YP82 F4WR43 A0A2A3EET7 E2APS4 A0A2Z5BRU1 A0A0L7QMM2 A0A0C9R5K1
PDB
1H76
E-value=6.08537e-08,
Score=139
Ontologies
Topology
SignalP
Position: 1 - 25,
Likelihood: 0.932134
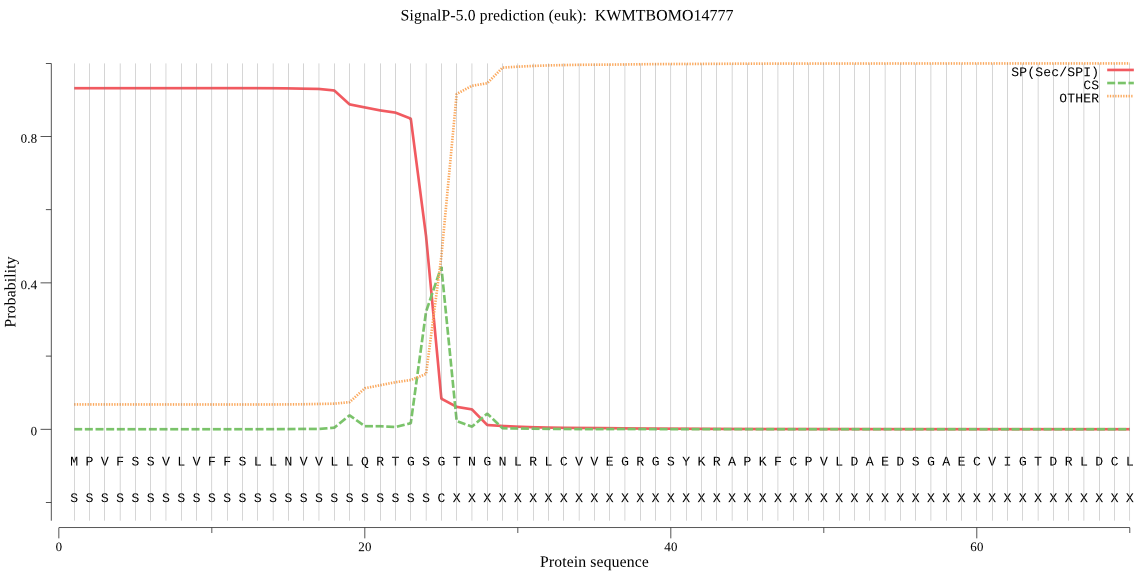
Length:
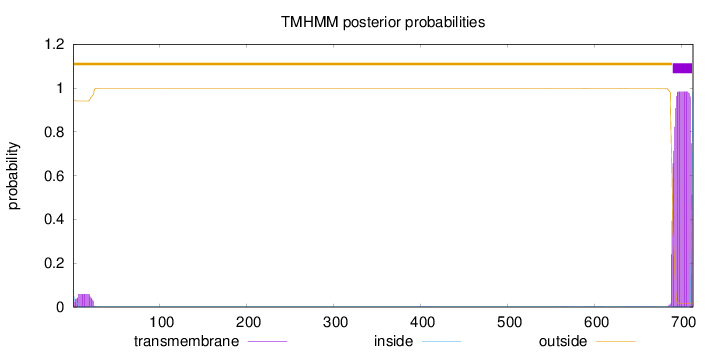
713
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.89407
Exp number, first 60 AAs:
1.14314
Total prob of N-in:
0.05859
outside
1 - 689
TMhelix
690 - 712
inside
713 - 713
Population Genetic Test Statistics
Pi
312.809384
Theta
167.319165
Tajima's D
2.479382
CLR
0.137705
CSRT
0.944102794860257
Interpretation
Uncertain
