Gene
KWMTBOMO14762
Pre Gene Modal
BGIBMGA003814
Annotation
PREDICTED:_glycerophosphodiester_phosphodiesterase_1-like_isoform_X1_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.293 PlasmaMembrane Reliability : 1.262
Sequence
CDS
ATGTTTTCGAAACTGTACACGATGTGCCAAATGTTTTGGAACAATATGTACTATTTATTTTCGTTTATAAACCTTTTAATTATTTATGTACATTTAGTTTTGCCCTTTGGTCTGGATGTCGGCTTGGCTTTTGCTGCTATGTATTACATCACCGCCCTGGAACGTCCTGATAAAGAGAGGGTACAGAAAATTTTTGGACACGATCCCTGGAGTAAAGATGAGGTTAAAAATTCTGAGGAAGTGGTGCATTGCATCGCTCACCGAGGAGCTGCACTTGACGCGCCGGAGAACACAATCGAAGCATTTAAATATTGCGTTGAACAAAACTGCAAGTTCGTCGAGATGGACGTGCGTTCGTTGCGAGACGGACAGCTGGTGCTGCTGCACGACGAGGGTCTGCAGCGTCTGACTGGAACCACGATCAACAACGTGCAGTGCGTCGACTGGGACGCCATAAAGGATTACAACGTGGCCGCCAATCATCCGAACAGGGAGCATTTCAAAGATGTGCGCCTATGTCTCTTCGAGGATGCACTGGACTATTTGCTCGAGAATAAATTTAAAATGATTATCGACGTCAAAGGCGTAGATCAAGAGGTGATATCCGGTGTCCTGGACGCGTTCGCCACCCGACCGGCTCTGTACGAGTCCGCAGCCGTGACCTGCTTCAACCCTTACCCGCTGTACCTGATCCGTAAAACCGATCCGAAAATCATTGGAGCTTTAAGCTATCGTCCGTATTGCTTTAGCGCGCAGGATTATGACGCCGAGAATGGACCAACGAATCCTAGGTTCCCTGACAGCGTCGTGCACCGGGTCGCGATGCGTGCGTGCGACGCGCTCCACGGGGTCGCGTGGCGGCGGCTCGCCCGCTGGTGCGGGGTCAGCGCCGTGCTCCTGCACAAGGACATTGTCAGCCCGAGCGAGGTGGAGTATTGGCGCGGGCTGGGCGTGCGGTGCGTCGGCTGGGCGGTGAACCGGCCCGTCGAGAAGCTGTACTGGCGCGGCGTGCTGAGGGCCGCCTACCTCGCGAGCACGCTGCTCGGCGAACCGGCCGTGGAGAAACAGAGCAAAGATAAGACCAGCTCTGACTGCGCAGCCTCTTCACCCGAACAAATGCTGGACCCGGAACAGACTGAGCAGAACGTATCGTCTGGACGAAATTAA
Protein
MFSKLYTMCQMFWNNMYYLFSFINLLIIYVHLVLPFGLDVGLAFAAMYYITALERPDKERVQKIFGHDPWSKDEVKNSEEVVHCIAHRGAALDAPENTIEAFKYCVEQNCKFVEMDVRSLRDGQLVLLHDEGLQRLTGTTINNVQCVDWDAIKDYNVAANHPNREHFKDVRLCLFEDALDYLLENKFKMIIDVKGVDQEVISGVLDAFATRPALYESAAVTCFNPYPLYLIRKTDPKIIGALSYRPYCFSAQDYDAENGPTNPRFPDSVVHRVAMRACDALHGVAWRRLARWCGVSAVLLHKDIVSPSEVEYWRGLGVRCVGWAVNRPVEKLYWRGVLRAAYLASTLLGEPAVEKQSKDKTSSDCAASSPEQMLDPEQTEQNVSSGRN
Summary
Similarity
Belongs to the amiloride-sensitive sodium channel (TC 1.A.6) family.
Uniprot
H9J2S7
A0A2A4JC31
A0A194Q0U6
A0A2Z5U775
A0A212F4U2
A0A2H1WNG8
+ More
A0A2W1BND0 A0A232ELT2 A0A0T6BC52 A0A067R7U7 A0A1B6LJD5 A0A1Y1N414 E2AT99 A0A1B6DZC9 A0A2J7PGR9 A0A026WZI2 A0A151IH07 E9IZ71 A0A0J7KYX9 E0V9S9 D6WBB2 A0A158NLD1 A0A154PTF7 A0A195B2C9 V5GN05 A0A0L7RIV3 A0A2A3EC29 A0A151XCC9 A0A146LD76 A0A195F2N9 A0A0N0U6W1 F4X6L8 K7JDI7 A0A2J7PGS8 A0A2P8ZBI1 A0A0A9X007 A0A195E6F0 A0A088AHI6 A0A1W4XD93 A0A3P8S7F3 A0A3Q1GJ84 A0A3Q1BFK7 A0A1V9XN02 A0A3B3I0F9 A8KBQ9 Q6P0H8 A0A3Q3CLX7 A0A3B4ZN46 A0A3P9JMQ6 A0A3P9L7L0 R4FLN1 A0A3P9MH18 T1HFM5 Q7ZUV3 A0A3P8Z4H1 A0A1D2NHR7 A0A3B4U3B0 A0A3B4XP93 A0A224XQU3 A0A2L2XW37 R7UV28 A0A023F6T3 A0A2L2XW40 A0A3Q1HX16 A0A3Q0S7W2 A0A3N0ZAH6 A0A3Q3SAY8 A0A2K5EEP4 H0X1U1 G1SK10 A0A3Q0SCU2 A0A250YFP8 A0A0S7GW15 A0A3B3QX43 A0A3B4U7M4 A0A3Q3XRW5 A0A3Q1JJV9
A0A2W1BND0 A0A232ELT2 A0A0T6BC52 A0A067R7U7 A0A1B6LJD5 A0A1Y1N414 E2AT99 A0A1B6DZC9 A0A2J7PGR9 A0A026WZI2 A0A151IH07 E9IZ71 A0A0J7KYX9 E0V9S9 D6WBB2 A0A158NLD1 A0A154PTF7 A0A195B2C9 V5GN05 A0A0L7RIV3 A0A2A3EC29 A0A151XCC9 A0A146LD76 A0A195F2N9 A0A0N0U6W1 F4X6L8 K7JDI7 A0A2J7PGS8 A0A2P8ZBI1 A0A0A9X007 A0A195E6F0 A0A088AHI6 A0A1W4XD93 A0A3P8S7F3 A0A3Q1GJ84 A0A3Q1BFK7 A0A1V9XN02 A0A3B3I0F9 A8KBQ9 Q6P0H8 A0A3Q3CLX7 A0A3B4ZN46 A0A3P9JMQ6 A0A3P9L7L0 R4FLN1 A0A3P9MH18 T1HFM5 Q7ZUV3 A0A3P8Z4H1 A0A1D2NHR7 A0A3B4U3B0 A0A3B4XP93 A0A224XQU3 A0A2L2XW37 R7UV28 A0A023F6T3 A0A2L2XW40 A0A3Q1HX16 A0A3Q0S7W2 A0A3N0ZAH6 A0A3Q3SAY8 A0A2K5EEP4 H0X1U1 G1SK10 A0A3Q0SCU2 A0A250YFP8 A0A0S7GW15 A0A3B3QX43 A0A3B4U7M4 A0A3Q3XRW5 A0A3Q1JJV9
Pubmed
EMBL
BABH01036940
BABH01036941
NWSH01002018
PCG69409.1
KQ459580
KPI99207.1
+ More
AP017492 BBB06765.1 AGBW02010304 OWR48765.1 ODYU01009894 SOQ54615.1 KZ150120 PZC73183.1 NNAY01003499 OXU19323.1 LJIG01002110 KRT84830.1 KK852879 KDR14441.1 GEBQ01016207 JAT23770.1 GEZM01015913 JAV91425.1 GL442545 EFN63344.1 GEDC01006484 GEDC01006278 JAS30814.1 JAS31020.1 NEVH01025164 PNF15533.1 KK107063 QOIP01000005 EZA61238.1 RLU22587.1 KQ977647 KYN00849.1 GL767119 EFZ14113.1 LBMM01001916 KMQ95551.1 DS234996 EEB10135.1 KQ971312 EEZ98717.1 ADTU01019320 KQ435119 KZC14714.1 KQ976662 KYM78435.1 GALX01002987 JAB65479.1 KQ414583 KOC70738.1 KZ288310 PBC28601.1 KQ982314 KYQ58015.1 GDHC01013030 JAQ05599.1 KQ981864 KYN34359.1 KQ435727 KOX77924.1 GL888818 EGI57800.1 AAZX01000022 AAZX01000037 AAZX01008686 AAZX01012922 AAZX01024815 PNF15532.1 PYGN01000115 PSN53858.1 GBHO01030598 JAG13006.1 KQ979568 KYN20780.1 MNPL01007199 OQR74890.1 BC154208 AAI54209.1 AL953841 BC065614 AAH65614.1 GAHY01001313 JAA76197.1 ACPB03012557 BC047818 AAH47818.1 LJIJ01000041 ODN04516.1 GFTR01005494 JAW10932.1 IAAA01002116 LAA00296.1 AMQN01006037 KB297448 ELU10493.1 GBBI01001621 JAC17091.1 IAAA01002114 IAAA01002115 LAA00292.1 RJVU01000282 ROL55436.1 AAQR03077450 AAQR03077451 AAQR03077452 AAQR03077453 AAQR03077454 AAGW02057074 GFFW01002343 JAV42445.1 GBYX01450982 GBYX01450980 JAO30522.1
AP017492 BBB06765.1 AGBW02010304 OWR48765.1 ODYU01009894 SOQ54615.1 KZ150120 PZC73183.1 NNAY01003499 OXU19323.1 LJIG01002110 KRT84830.1 KK852879 KDR14441.1 GEBQ01016207 JAT23770.1 GEZM01015913 JAV91425.1 GL442545 EFN63344.1 GEDC01006484 GEDC01006278 JAS30814.1 JAS31020.1 NEVH01025164 PNF15533.1 KK107063 QOIP01000005 EZA61238.1 RLU22587.1 KQ977647 KYN00849.1 GL767119 EFZ14113.1 LBMM01001916 KMQ95551.1 DS234996 EEB10135.1 KQ971312 EEZ98717.1 ADTU01019320 KQ435119 KZC14714.1 KQ976662 KYM78435.1 GALX01002987 JAB65479.1 KQ414583 KOC70738.1 KZ288310 PBC28601.1 KQ982314 KYQ58015.1 GDHC01013030 JAQ05599.1 KQ981864 KYN34359.1 KQ435727 KOX77924.1 GL888818 EGI57800.1 AAZX01000022 AAZX01000037 AAZX01008686 AAZX01012922 AAZX01024815 PNF15532.1 PYGN01000115 PSN53858.1 GBHO01030598 JAG13006.1 KQ979568 KYN20780.1 MNPL01007199 OQR74890.1 BC154208 AAI54209.1 AL953841 BC065614 AAH65614.1 GAHY01001313 JAA76197.1 ACPB03012557 BC047818 AAH47818.1 LJIJ01000041 ODN04516.1 GFTR01005494 JAW10932.1 IAAA01002116 LAA00296.1 AMQN01006037 KB297448 ELU10493.1 GBBI01001621 JAC17091.1 IAAA01002114 IAAA01002115 LAA00292.1 RJVU01000282 ROL55436.1 AAQR03077450 AAQR03077451 AAQR03077452 AAQR03077453 AAQR03077454 AAGW02057074 GFFW01002343 JAV42445.1 GBYX01450982 GBYX01450980 JAO30522.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000215335
UP000027135
+ More
UP000000311 UP000235965 UP000053097 UP000279307 UP000078542 UP000036403 UP000009046 UP000007266 UP000005205 UP000076502 UP000078540 UP000053825 UP000242457 UP000075809 UP000078541 UP000053105 UP000007755 UP000002358 UP000245037 UP000078492 UP000005203 UP000192223 UP000265080 UP000257200 UP000257160 UP000192247 UP000001038 UP000000437 UP000264840 UP000261400 UP000265200 UP000265180 UP000015103 UP000265140 UP000094527 UP000261420 UP000261360 UP000014760 UP000265040 UP000261340 UP000261640 UP000233020 UP000005225 UP000001811 UP000261540 UP000261620
UP000000311 UP000235965 UP000053097 UP000279307 UP000078542 UP000036403 UP000009046 UP000007266 UP000005205 UP000076502 UP000078540 UP000053825 UP000242457 UP000075809 UP000078541 UP000053105 UP000007755 UP000002358 UP000245037 UP000078492 UP000005203 UP000192223 UP000265080 UP000257200 UP000257160 UP000192247 UP000001038 UP000000437 UP000264840 UP000261400 UP000265200 UP000265180 UP000015103 UP000265140 UP000094527 UP000261420 UP000261360 UP000014760 UP000265040 UP000261340 UP000261640 UP000233020 UP000005225 UP000001811 UP000261540 UP000261620
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
H9J2S7
A0A2A4JC31
A0A194Q0U6
A0A2Z5U775
A0A212F4U2
A0A2H1WNG8
+ More
A0A2W1BND0 A0A232ELT2 A0A0T6BC52 A0A067R7U7 A0A1B6LJD5 A0A1Y1N414 E2AT99 A0A1B6DZC9 A0A2J7PGR9 A0A026WZI2 A0A151IH07 E9IZ71 A0A0J7KYX9 E0V9S9 D6WBB2 A0A158NLD1 A0A154PTF7 A0A195B2C9 V5GN05 A0A0L7RIV3 A0A2A3EC29 A0A151XCC9 A0A146LD76 A0A195F2N9 A0A0N0U6W1 F4X6L8 K7JDI7 A0A2J7PGS8 A0A2P8ZBI1 A0A0A9X007 A0A195E6F0 A0A088AHI6 A0A1W4XD93 A0A3P8S7F3 A0A3Q1GJ84 A0A3Q1BFK7 A0A1V9XN02 A0A3B3I0F9 A8KBQ9 Q6P0H8 A0A3Q3CLX7 A0A3B4ZN46 A0A3P9JMQ6 A0A3P9L7L0 R4FLN1 A0A3P9MH18 T1HFM5 Q7ZUV3 A0A3P8Z4H1 A0A1D2NHR7 A0A3B4U3B0 A0A3B4XP93 A0A224XQU3 A0A2L2XW37 R7UV28 A0A023F6T3 A0A2L2XW40 A0A3Q1HX16 A0A3Q0S7W2 A0A3N0ZAH6 A0A3Q3SAY8 A0A2K5EEP4 H0X1U1 G1SK10 A0A3Q0SCU2 A0A250YFP8 A0A0S7GW15 A0A3B3QX43 A0A3B4U7M4 A0A3Q3XRW5 A0A3Q1JJV9
A0A2W1BND0 A0A232ELT2 A0A0T6BC52 A0A067R7U7 A0A1B6LJD5 A0A1Y1N414 E2AT99 A0A1B6DZC9 A0A2J7PGR9 A0A026WZI2 A0A151IH07 E9IZ71 A0A0J7KYX9 E0V9S9 D6WBB2 A0A158NLD1 A0A154PTF7 A0A195B2C9 V5GN05 A0A0L7RIV3 A0A2A3EC29 A0A151XCC9 A0A146LD76 A0A195F2N9 A0A0N0U6W1 F4X6L8 K7JDI7 A0A2J7PGS8 A0A2P8ZBI1 A0A0A9X007 A0A195E6F0 A0A088AHI6 A0A1W4XD93 A0A3P8S7F3 A0A3Q1GJ84 A0A3Q1BFK7 A0A1V9XN02 A0A3B3I0F9 A8KBQ9 Q6P0H8 A0A3Q3CLX7 A0A3B4ZN46 A0A3P9JMQ6 A0A3P9L7L0 R4FLN1 A0A3P9MH18 T1HFM5 Q7ZUV3 A0A3P8Z4H1 A0A1D2NHR7 A0A3B4U3B0 A0A3B4XP93 A0A224XQU3 A0A2L2XW37 R7UV28 A0A023F6T3 A0A2L2XW40 A0A3Q1HX16 A0A3Q0S7W2 A0A3N0ZAH6 A0A3Q3SAY8 A0A2K5EEP4 H0X1U1 G1SK10 A0A3Q0SCU2 A0A250YFP8 A0A0S7GW15 A0A3B3QX43 A0A3B4U7M4 A0A3Q3XRW5 A0A3Q1JJV9
PDB
2PZ0
E-value=5.48036e-10,
Score=154
Ontologies
GO
PANTHER
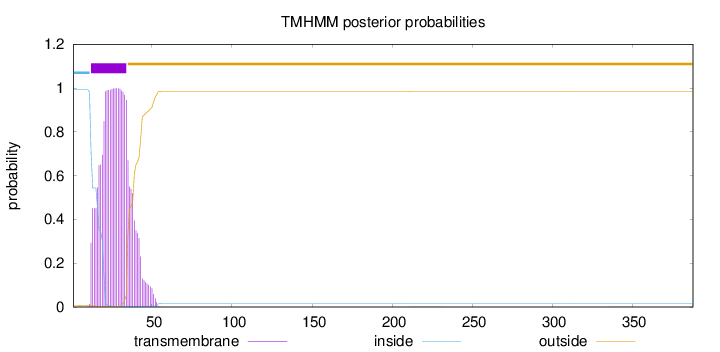
Topology
Length:
388
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.67177
Exp number, first 60 AAs:
23.6605
Total prob of N-in:
0.99230
POSSIBLE N-term signal
sequence
inside
1 - 11
TMhelix
12 - 34
outside
35 - 388
Population Genetic Test Statistics
Pi
52.102499
Theta
51.139205
Tajima's D
0.162814
CLR
116.267681
CSRT
0.417979101044948
Interpretation
Possibly Positive selection
