Gene
KWMTBOMO14761 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003815
Annotation
PREDICTED:_dihydrolipoyllysine-residue_succinyltransferase_component_of_2-oxoglutarate_dehydrogenase_complex?_mitochondrial-like_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.949
Sequence
CDS
ATGCTACGACGCTGCTCGAAGCACATCCAGACGCTGTACCGGCGTCAGGGCCAGTCCATCCGCTTCAAGTCAACAACTCAAACGCCCAAAATACTTGCACCCTTACACGCAACCAAACTGAACCAACCGAGAGCTCTGGTCGCCCACAATCAAGTAGCATCCATCCACTTCACGAATCCCCTGCTTGTAGAGCAAGATGTGACCACTCCAAGTTTCCCGGACTCCGTTTCTGAAGGTGACGTCAAATTAGATAAGAAAGTGGGAGACAGCGTAGCCGCCGATGAAGTGGTCCTGGAAATCGAGACGGACAAAACCGCCATTCCGGTCATGGCGCCCGGCCATGGTGTCATCAAGGAGTTGTACGTCAAAGACGGAGAAACCGTTAAAGCTGGACAAAAGCTGTTTAGGTTAGAGATCACAGGGGAAGCACCTAAAAAGGCTGCAGCTACTGAGGCTCCGAAGGTTGATACACCACCGCCGCCGCCGCCGCCTCCTCAAGCTGCCGCTCCGCCTCCGCCCCCACCTCCCGCCGCGGCAGCTCCTCCCTCACCACCCCCACCTACCCCTAAAGCCGCTCCGCCTCCGCCCGCTGCTCCCATCTCCTCAATACCAGTCGCCGCCATCCGTCACGCTCAGGCGATCGAGACAGCAACTGTTAAAGTTCCTCCACAGGACTACAGCAAAGAGATCGCCGGCACGCGCACCGAGCAACGGGTCAAGATGAACAGGATGAGGCAACGCATCGCTCAGCGTTTGAAGGACGCTCAGAACACGAACGCCATGTTGACGACATTCAACGAGATCGACATGTCCCACATCATGGCATTCCGTAAGAAGCACCTCGACACCTTCACTAAGAAGCACAGCATCAAACTCGGTCTGATGTCACCTTTCGTGAAGGCGGCCGCCAACGCCCTGATGGACCAGCCCGTGGTGAACGCTGTTATCGAAGAAAACGAGATAATTTATAGGGACTACGTAGATATTTCGGTAGCGGTCGCCACACCGAAAGGTCTCGTCGTGCCGGTCATCAGAAACGTTCAGAACATGACTTATGCTGATATCGAGCTCACCATAGCTGGGCTAGCGGAAAAAGCGAGAACCGGTAAGCTCACGATCGAAGAAATGGACGGCGGAACGTTCACCATCAGCAATGGCGGCGTTTTCGGGTCTCTGATGGGAACGCCAATCATCAATCCGCCTCAATCTGCTATACTGGGTATGCACGGCATCTTCGAGCGGCCCATCGCTCTGAACGGCCAAGTCGTGATCAGGCCCATGATGTATATTGCCCTAACTTACGACCACAGGCTGATAGACGGCCGCGAAGCTGTACTCTTCCTCAGAAAGATCAAGGAAGGCGTCGAAGATCCCGCCACTATCGTCGCTGGCTTGTAA
Protein
MLRRCSKHIQTLYRRQGQSIRFKSTTQTPKILAPLHATKLNQPRALVAHNQVASIHFTNPLLVEQDVTTPSFPDSVSEGDVKLDKKVGDSVAADEVVLEIETDKTAIPVMAPGHGVIKELYVKDGETVKAGQKLFRLEITGEAPKKAAATEAPKVDTPPPPPPPPQAAAPPPPPPPAAAAPPSPPPPTPKAAPPPPAAPISSIPVAAIRHAQAIETATVKVPPQDYSKEIAGTRTEQRVKMNRMRQRIAQRLKDAQNTNAMLTTFNEIDMSHIMAFRKKHLDTFTKKHSIKLGLMSPFVKAAANALMDQPVVNAVIEENEIIYRDYVDISVAVATPKGLVVPVIRNVQNMTYADIELTIAGLAEKARTGKLTIEEMDGGTFTISNGGVFGSLMGTPIINPPQSAILGMHGIFERPIALNGQVVIRPMMYIALTYDHRLIDGREAVLFLRKIKEGVEDPATIVAGL
Summary
Uniprot
EMBL
Proteomes
Interpro
SUPFAM
SSF51230
SSF51230
Gene 3D
ProteinModelPortal
PDB
1SCZ
E-value=1.46271e-74,
Score=712
Ontologies
PATHWAY
00020
Citrate cycle (TCA cycle) - Bombyx mori (domestic silkworm)
00310 Lysine degradation - Bombyx mori (domestic silkworm)
00380 Tryptophan metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
00310 Lysine degradation - Bombyx mori (domestic silkworm)
00380 Tryptophan metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
GO
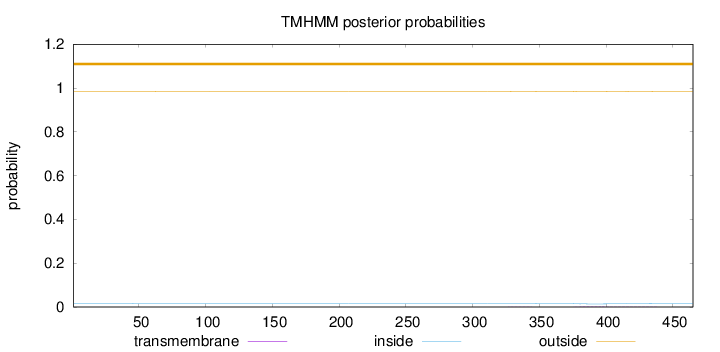
Topology
Length:
465
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.07227
Exp number, first 60 AAs:
0.00149
Total prob of N-in:
0.01526
outside
1 - 465
Population Genetic Test Statistics
Pi
177.978688
Theta
151.360896
Tajima's D
0.73893
CLR
0.285194
CSRT
0.585570721463927
Interpretation
Uncertain
