Pre Gene Modal
BGIBMGA003817
Annotation
UDP-glycosyltransferase_UGT41A1_precursor_[Bombyx_mori]
Full name
UDP-glucuronosyltransferase
Location in the cell
PlasmaMembrane Reliability : 3.181
Sequence
CDS
ATGAGGTGTCTCGGACTGCTTTTCTTCTTGGTGTGCGTGGTGACGTCGGCGCGTGCGTACCACGTGCTCTGCGTGTTCCCGATACCTTCGCGCAGCCACAACTCCCTCGGCAAAGGGATAGTCGACGCGTTGCTTGAGGCTGGTCACGAGGTGACCTGGGTGACTCCGTATCCTCCGAGCGAACTGGCCAAAGGTCTGAAGATAGTCGACGTCAGCGCTACAGTATCTATAAGTAAAACGGTGGACATGCATGAGCAGAGAAACTCGAATACTGGTGTATCTTTCGTGAAGGCTTTGGCCGAGAACATCACCAGGGTTTCCCTGGCAACGCCGGCTCTACAGCAGGCGATCGTCCAGGGGAAATACGACGCGGTCATCACGGAAACGTTCTTCAACGATGCCGAAGCTGGTTACGGGGCAGTACTTCAAGTGCCCTGGATCTTGATGTCCAGCATCGCAATGATGCCTCAGTTGGAAGCCATCGTCGATGAGGTGCGGTCGGTCACCACCATACCGCTGCTGTTCAACAACGCCCCAACCCCCATGGGTTTCTGGGACAGACTGAAGAATGTCTTTCTCCACAGCGTCATGGTGATATCTGATTGGCTAGACCGCCCCAAGACGGTGGCATTCTACGAGTCCCTATTCGCGCCGCTGGCCACCGCCCGGGGGGTCGCGCTGCCCCCCTTCGAGGAAGCCCTGTACAACGTGTCGGTGCTGCTTGTGAACTCCCACCCGGCCTTCGCTCCGCCCCTGAGCTTGCCCCCGAACGTCGTCGAGATCGCGGGCTACCATATCGACCCAAAAACACCCCCCTTGCCGAAGGACCTGCAGAGCATCCTGGACTCGTCGCCGCAGGGCGTGGTGTACTTCAGCATGGGCTCGGTGCTCAAGTCGTCCAAACTCTCCGAACAAACTAGACGAGAGTTATTGGATGTGTTTGGGTCTATACCGCAGACTGTCCTTTGGAAGTTCGAAGAGGATCTCCAAGATCTACCGAAGAACGTTCACATTAGATCGTGGATGCCTCAGTCCAGTATACTAGCTCATCCGAACATGAAGGTCTTCATCACGCACGGCGGCTTACTCAGCATCCTCGAGACCTTGCATTACGGAGTACCGATCCTCGCTGTGCCGGTGTTCGGGGACCAGCCCTCAAACGCCAATAGCGCTGTCCGTAACGGATTTGCGAAAAGTATAGAGTACAAGCCTGATATGGCTAAGGACATGAAGGTGGCCCTGAATGAAATGCTGAGCGACGACAGTTACTACAAACGTGCGAGATATCTGTCGAAGATCTTCGGCGATAAGTTGGTGCCGCCGGCCAAGGTGATATCGCATTATGTGAAGGTCGCCATCGAAACTAATGGAGCCTACCATCTGCGCTCTAAGAGCCTGCTGTATCCGTGGTACCAGCGTTGGCTCGTGGACATCATAGCGGCACTACTTCTGGCGTGCCTCGCGGTGTACGTAGTGGCCAGGAGGGTCTTGTGTTACCTCTACACCTCCGTGACCGGTGGGGGCTGTAACAGGAGTGTTAAAGTCAAGAAAAACTAA
Protein
MRCLGLLFFLVCVVTSARAYHVLCVFPIPSRSHNSLGKGIVDALLEAGHEVTWVTPYPPSELAKGLKIVDVSATVSISKTVDMHEQRNSNTGVSFVKALAENITRVSLATPALQQAIVQGKYDAVITETFFNDAEAGYGAVLQVPWILMSSIAMMPQLEAIVDEVRSVTTIPLLFNNAPTPMGFWDRLKNVFLHSVMVISDWLDRPKTVAFYESLFAPLATARGVALPPFEEALYNVSVLLVNSHPAFAPPLSLPPNVVEIAGYHIDPKTPPLPKDLQSILDSSPQGVVYFSMGSVLKSSKLSEQTRRELLDVFGSIPQTVLWKFEEDLQDLPKNVHIRSWMPQSSILAHPNMKVFITHGGLLSILETLHYGVPILAVPVFGDQPSNANSAVRNGFAKSIEYKPDMAKDMKVALNEMLSDDSYYKRARYLSKIFGDKLVPPAKVISHYVKVAIETNGAYHLRSKSLLYPWYQRWLVDIIAALLLACLAVYVVARRVLCYLYTSVTGGGCNRSVKVKKN
Summary
Catalytic Activity
glucuronate acceptor + UDP-alpha-D-glucuronate = acceptor beta-D-glucuronoside + H(+) + UDP
Similarity
Belongs to the UDP-glycosyltransferase family.
Feature
chain UDP-glucuronosyltransferase
Uniprot
G9LPV2
G9LPV3
G9LPV4
H9J2T0
H9IVQ7
H9J2U8
+ More
G9LPR1 A0A2W1BAL9 A0A024AGL8 A0A2A4JV21 A0A2W1BD64 A0A0N0PBY8 A0A2A4JD43 A0A140EA38 A0A1L8D6F2 G9LPQ8 A0A2H1VNG6 A0A286MXP3 A0A2H4WB50 G9LPR0 A0A2W1BBU4 A0A140EA39 A0A212EW66 A0A2A4JUA9 A0A2A4JVG7 G9LPQ9 A0A2W1BEH1 A0A2H4WB58 A0A2H4WB67 A0A2A4J0B4 A0A191T1M8 A0A191T1M6 A0A2A4J6T0 G9LPQ4 A0A3S2NJP8 A0A2W1BQJ6 A0A194RDA8 A0A194Q0G5 A0A2H4WB53 G9LPQ6 A0A2H1VVH6 A0A140EA37 G9LPU4 A0A191T2G6 Q8WPG4 A0A024AGG4 G9LPU0 A0A024AG04 G9LPQ3 G9LPQ0 G9LPU3 A0A2H1WNF9 A0A191T2K0 D6RUU7 A0A1L8D6H0 A0A024AH17 A0A2W1BXG7 A0A191T2K2 A0A2W1BQJ5 A0A2A4J325 A0A191T2J8 A0A1L8D6E4 D6RUU6 G9LPQ5 A0A2W1BVZ0 G9LPU8 A0A2A4K2B6 A0A2H1V1W5 G9LPU6 A0A2A4J419 A0A2W1BX32 A0A140EA36 A0A212FKT5 A0A212FD09 D6RUV0 A0A024AHN0 G9LPU7 A0A2H4WB78 G9LPQ1 A0A194RKA9 A0A191T1P1 A0A2W1BQS6 I4DN89 A0A194RKU4 A0A0N1IN21 A0A2A4K0I8 D6RUV1 G9LPQ7 A0A2H1X1Y8 A0A3S2LTQ3 A0A3S2LD93 H9JL88 A0A194R8C7 A0A191T1N5 A0A1L8D6R9 I4DR17
G9LPR1 A0A2W1BAL9 A0A024AGL8 A0A2A4JV21 A0A2W1BD64 A0A0N0PBY8 A0A2A4JD43 A0A140EA38 A0A1L8D6F2 G9LPQ8 A0A2H1VNG6 A0A286MXP3 A0A2H4WB50 G9LPR0 A0A2W1BBU4 A0A140EA39 A0A212EW66 A0A2A4JUA9 A0A2A4JVG7 G9LPQ9 A0A2W1BEH1 A0A2H4WB58 A0A2H4WB67 A0A2A4J0B4 A0A191T1M8 A0A191T1M6 A0A2A4J6T0 G9LPQ4 A0A3S2NJP8 A0A2W1BQJ6 A0A194RDA8 A0A194Q0G5 A0A2H4WB53 G9LPQ6 A0A2H1VVH6 A0A140EA37 G9LPU4 A0A191T2G6 Q8WPG4 A0A024AGG4 G9LPU0 A0A024AG04 G9LPQ3 G9LPQ0 G9LPU3 A0A2H1WNF9 A0A191T2K0 D6RUU7 A0A1L8D6H0 A0A024AH17 A0A2W1BXG7 A0A191T2K2 A0A2W1BQJ5 A0A2A4J325 A0A191T2J8 A0A1L8D6E4 D6RUU6 G9LPQ5 A0A2W1BVZ0 G9LPU8 A0A2A4K2B6 A0A2H1V1W5 G9LPU6 A0A2A4J419 A0A2W1BX32 A0A140EA36 A0A212FKT5 A0A212FD09 D6RUV0 A0A024AHN0 G9LPU7 A0A2H4WB78 G9LPQ1 A0A194RKA9 A0A191T1P1 A0A2W1BQS6 I4DN89 A0A194RKU4 A0A0N1IN21 A0A2A4K0I8 D6RUV1 G9LPQ7 A0A2H1X1Y8 A0A3S2LTQ3 A0A3S2LD93 H9JL88 A0A194R8C7 A0A191T1N5 A0A1L8D6R9 I4DR17
EC Number
2.4.1.17
Pubmed
EMBL
JQ070259
AEW43175.1
JQ070260
AEW43176.1
JQ070261
AEW43177.1
+ More
BABH01036935 BABH01036936 BABH01036937 BABH01042490 BABH01036929 BABH01036930 JQ070218 AEW43134.1 KZ150343 PZC71295.1 KF777117 AHY99688.1 NWSH01000566 PCG75616.1 KZ150120 PZC73179.1 KQ460735 KPJ12633.1 NWSH01001859 PCG69891.1 KU214509 AMK97475.1 GEYN01000097 JAV02032.1 JQ070215 AEW43131.1 ODYU01003485 SOQ42346.1 MF684361 ASX93995.1 KY202927 AUC64271.1 JQ070217 AEW43133.1 PZC71294.1 KU214510 AMK97476.1 AGBW02012074 OWR45723.1 PCG75617.1 PCG75614.1 JQ070216 AEW43132.1 PZC71296.1 KY202932 AUC64276.1 KY202937 AUC64281.1 NWSH01003979 PCG65597.1 KU680303 ANI22009.1 KU680302 ANI22008.1 NWSH01002698 PCG67661.1 JQ070211 AEW43127.1 RSAL01000067 RVE49308.1 KZ149930 PZC77372.1 KQ460597 KPJ13896.1 KQ459582 KPI98793.1 KY202925 AUC64269.1 JQ070213 AEW43129.1 ODYU01004685 SOQ44830.1 KU214508 AMK97474.1 JQ070251 AEW43167.1 KU680295 ANI22001.1 AF324465 AAL55994.1 KF777115 AHY99686.1 JQ070247 AEW43163.1 KF777114 AHY99685.1 JQ070210 AEW43126.1 JQ070207 AEW43123.1 JQ070250 AEW43166.1 ODYU01009829 SOQ54516.1 KU680299 ANI22005.1 JQ070252 AB539964 AEW43168.1 BAJ08154.1 GEYN01000076 JAV02053.1 KF777116 AHY99687.1 PZC77366.1 KU680304 ANI22010.1 PZC77369.1 NWSH01003818 PCG65793.1 KU680294 ANI22000.1 GEYN01000100 JAV02029.1 AB539963 BAJ08153.1 JQ070212 AEW43128.1 PZC77367.1 JQ070255 AEW43171.1 NWSH01000282 PCG77782.1 ODYU01000116 SOQ34342.1 JQ070253 AEW43169.1 NWSH01003535 PCG66153.1 PZC77370.1 KU214507 AMK97473.1 AGBW02007992 OWR54346.1 AGBW02009118 OWR51610.1 AB539967 BAJ08157.1 KF777113 AHY99684.1 JQ070254 AEW43170.1 KY202946 AUC64290.1 JQ070208 AEW43124.1 KQ460075 KPJ17977.1 KU680301 ANI22007.1 PZC77368.1 AK402777 BAM19379.1 KPJ17950.1 KQ459356 KPJ01283.1 PCG77781.1 JQ070258 AB539968 AEW43174.1 BAJ08158.1 JQ070214 AEW43130.1 ODYU01012744 SOQ59206.1 RSAL01000210 RVE44215.1 RVE44216.1 BABH01011785 BABH01011786 BABH01011787 KPJ13897.1 KU680296 ANI22002.1 GEYN01000085 JAV02044.1 AK404892 BAM20357.1
BABH01036935 BABH01036936 BABH01036937 BABH01042490 BABH01036929 BABH01036930 JQ070218 AEW43134.1 KZ150343 PZC71295.1 KF777117 AHY99688.1 NWSH01000566 PCG75616.1 KZ150120 PZC73179.1 KQ460735 KPJ12633.1 NWSH01001859 PCG69891.1 KU214509 AMK97475.1 GEYN01000097 JAV02032.1 JQ070215 AEW43131.1 ODYU01003485 SOQ42346.1 MF684361 ASX93995.1 KY202927 AUC64271.1 JQ070217 AEW43133.1 PZC71294.1 KU214510 AMK97476.1 AGBW02012074 OWR45723.1 PCG75617.1 PCG75614.1 JQ070216 AEW43132.1 PZC71296.1 KY202932 AUC64276.1 KY202937 AUC64281.1 NWSH01003979 PCG65597.1 KU680303 ANI22009.1 KU680302 ANI22008.1 NWSH01002698 PCG67661.1 JQ070211 AEW43127.1 RSAL01000067 RVE49308.1 KZ149930 PZC77372.1 KQ460597 KPJ13896.1 KQ459582 KPI98793.1 KY202925 AUC64269.1 JQ070213 AEW43129.1 ODYU01004685 SOQ44830.1 KU214508 AMK97474.1 JQ070251 AEW43167.1 KU680295 ANI22001.1 AF324465 AAL55994.1 KF777115 AHY99686.1 JQ070247 AEW43163.1 KF777114 AHY99685.1 JQ070210 AEW43126.1 JQ070207 AEW43123.1 JQ070250 AEW43166.1 ODYU01009829 SOQ54516.1 KU680299 ANI22005.1 JQ070252 AB539964 AEW43168.1 BAJ08154.1 GEYN01000076 JAV02053.1 KF777116 AHY99687.1 PZC77366.1 KU680304 ANI22010.1 PZC77369.1 NWSH01003818 PCG65793.1 KU680294 ANI22000.1 GEYN01000100 JAV02029.1 AB539963 BAJ08153.1 JQ070212 AEW43128.1 PZC77367.1 JQ070255 AEW43171.1 NWSH01000282 PCG77782.1 ODYU01000116 SOQ34342.1 JQ070253 AEW43169.1 NWSH01003535 PCG66153.1 PZC77370.1 KU214507 AMK97473.1 AGBW02007992 OWR54346.1 AGBW02009118 OWR51610.1 AB539967 BAJ08157.1 KF777113 AHY99684.1 JQ070254 AEW43170.1 KY202946 AUC64290.1 JQ070208 AEW43124.1 KQ460075 KPJ17977.1 KU680301 ANI22007.1 PZC77368.1 AK402777 BAM19379.1 KPJ17950.1 KQ459356 KPJ01283.1 PCG77781.1 JQ070258 AB539968 AEW43174.1 BAJ08158.1 JQ070214 AEW43130.1 ODYU01012744 SOQ59206.1 RSAL01000210 RVE44215.1 RVE44216.1 BABH01011785 BABH01011786 BABH01011787 KPJ13897.1 KU680296 ANI22002.1 GEYN01000085 JAV02044.1 AK404892 BAM20357.1
Proteomes
PRIDE
Pfam
PF00201 UDPGT
ProteinModelPortal
G9LPV2
G9LPV3
G9LPV4
H9J2T0
H9IVQ7
H9J2U8
+ More
G9LPR1 A0A2W1BAL9 A0A024AGL8 A0A2A4JV21 A0A2W1BD64 A0A0N0PBY8 A0A2A4JD43 A0A140EA38 A0A1L8D6F2 G9LPQ8 A0A2H1VNG6 A0A286MXP3 A0A2H4WB50 G9LPR0 A0A2W1BBU4 A0A140EA39 A0A212EW66 A0A2A4JUA9 A0A2A4JVG7 G9LPQ9 A0A2W1BEH1 A0A2H4WB58 A0A2H4WB67 A0A2A4J0B4 A0A191T1M8 A0A191T1M6 A0A2A4J6T0 G9LPQ4 A0A3S2NJP8 A0A2W1BQJ6 A0A194RDA8 A0A194Q0G5 A0A2H4WB53 G9LPQ6 A0A2H1VVH6 A0A140EA37 G9LPU4 A0A191T2G6 Q8WPG4 A0A024AGG4 G9LPU0 A0A024AG04 G9LPQ3 G9LPQ0 G9LPU3 A0A2H1WNF9 A0A191T2K0 D6RUU7 A0A1L8D6H0 A0A024AH17 A0A2W1BXG7 A0A191T2K2 A0A2W1BQJ5 A0A2A4J325 A0A191T2J8 A0A1L8D6E4 D6RUU6 G9LPQ5 A0A2W1BVZ0 G9LPU8 A0A2A4K2B6 A0A2H1V1W5 G9LPU6 A0A2A4J419 A0A2W1BX32 A0A140EA36 A0A212FKT5 A0A212FD09 D6RUV0 A0A024AHN0 G9LPU7 A0A2H4WB78 G9LPQ1 A0A194RKA9 A0A191T1P1 A0A2W1BQS6 I4DN89 A0A194RKU4 A0A0N1IN21 A0A2A4K0I8 D6RUV1 G9LPQ7 A0A2H1X1Y8 A0A3S2LTQ3 A0A3S2LD93 H9JL88 A0A194R8C7 A0A191T1N5 A0A1L8D6R9 I4DR17
G9LPR1 A0A2W1BAL9 A0A024AGL8 A0A2A4JV21 A0A2W1BD64 A0A0N0PBY8 A0A2A4JD43 A0A140EA38 A0A1L8D6F2 G9LPQ8 A0A2H1VNG6 A0A286MXP3 A0A2H4WB50 G9LPR0 A0A2W1BBU4 A0A140EA39 A0A212EW66 A0A2A4JUA9 A0A2A4JVG7 G9LPQ9 A0A2W1BEH1 A0A2H4WB58 A0A2H4WB67 A0A2A4J0B4 A0A191T1M8 A0A191T1M6 A0A2A4J6T0 G9LPQ4 A0A3S2NJP8 A0A2W1BQJ6 A0A194RDA8 A0A194Q0G5 A0A2H4WB53 G9LPQ6 A0A2H1VVH6 A0A140EA37 G9LPU4 A0A191T2G6 Q8WPG4 A0A024AGG4 G9LPU0 A0A024AG04 G9LPQ3 G9LPQ0 G9LPU3 A0A2H1WNF9 A0A191T2K0 D6RUU7 A0A1L8D6H0 A0A024AH17 A0A2W1BXG7 A0A191T2K2 A0A2W1BQJ5 A0A2A4J325 A0A191T2J8 A0A1L8D6E4 D6RUU6 G9LPQ5 A0A2W1BVZ0 G9LPU8 A0A2A4K2B6 A0A2H1V1W5 G9LPU6 A0A2A4J419 A0A2W1BX32 A0A140EA36 A0A212FKT5 A0A212FD09 D6RUV0 A0A024AHN0 G9LPU7 A0A2H4WB78 G9LPQ1 A0A194RKA9 A0A191T1P1 A0A2W1BQS6 I4DN89 A0A194RKU4 A0A0N1IN21 A0A2A4K0I8 D6RUV1 G9LPQ7 A0A2H1X1Y8 A0A3S2LTQ3 A0A3S2LD93 H9JL88 A0A194R8C7 A0A191T1N5 A0A1L8D6R9 I4DR17
PDB
2O6L
E-value=4.95824e-25,
Score=285
Ontologies
PATHWAY
00040
Pentose and glucuronate interconversions - Bombyx mori (domestic silkworm)
00053 Ascorbate and aldarate metabolism - Bombyx mori (domestic silkworm)
00830 Retinol metabolism - Bombyx mori (domestic silkworm)
00860 Porphyrin and chlorophyll metabolism - Bombyx mori (domestic silkworm)
00980 Metabolism of xenobiotics by cytochrome P450 - Bombyx mori (domestic silkworm)
00982 Drug metabolism - cytochrome P450 - Bombyx mori (domestic silkworm)
00983 Drug metabolism - other enzymes - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
00053 Ascorbate and aldarate metabolism - Bombyx mori (domestic silkworm)
00830 Retinol metabolism - Bombyx mori (domestic silkworm)
00860 Porphyrin and chlorophyll metabolism - Bombyx mori (domestic silkworm)
00980 Metabolism of xenobiotics by cytochrome P450 - Bombyx mori (domestic silkworm)
00982 Drug metabolism - cytochrome P450 - Bombyx mori (domestic silkworm)
00983 Drug metabolism - other enzymes - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
GO
Topology
Subcellular location
Membrane
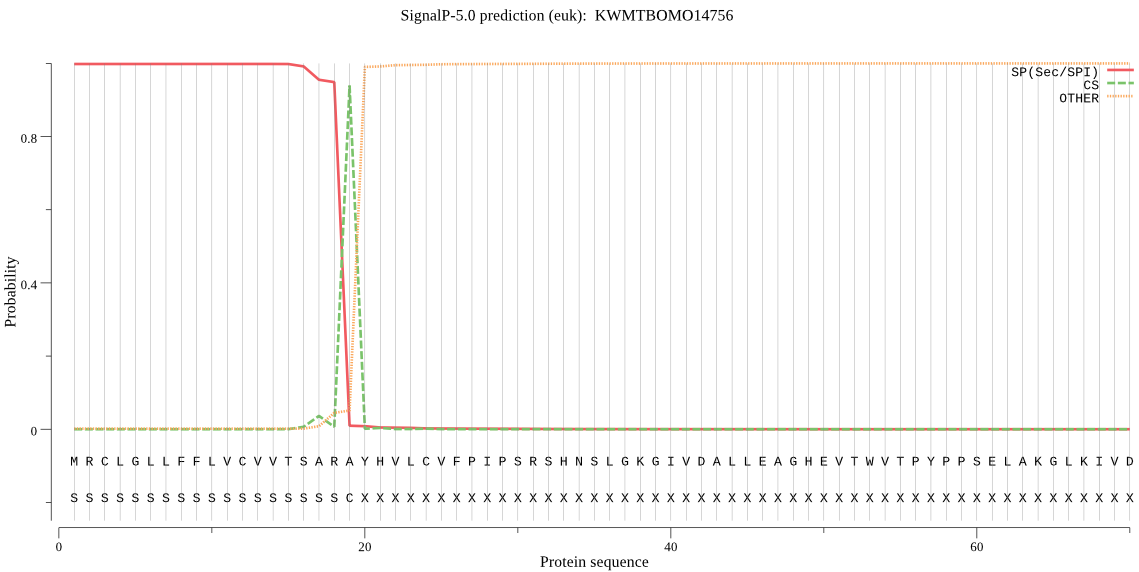
SignalP
Position: 1 - 19,
Likelihood: 0.997984
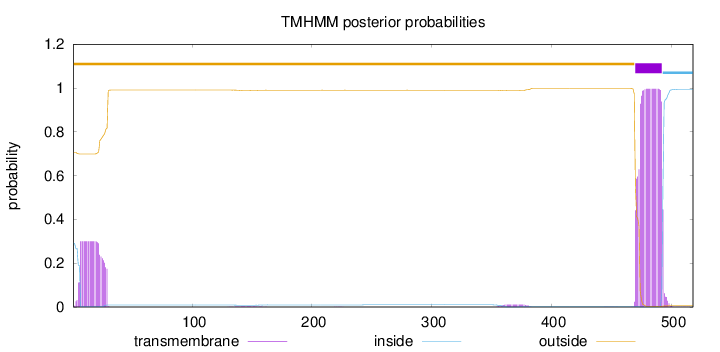
Length:
518
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
28.5335
Exp number, first 60 AAs:
6.53421999999999
Total prob of N-in:
0.29319
outside
1 - 469
TMhelix
470 - 492
inside
493 - 518
Population Genetic Test Statistics
Pi
208.642853
Theta
170.601519
Tajima's D
0.512248
CLR
1.432403
CSRT
0.517774111294435
Interpretation
Uncertain
