Gene
KWMTBOMO14752 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003833
Annotation
PREDICTED:_H/ACA_ribonucleoprotein_complex_subunit_4_[Amyelois_transitella]
Full name
H/ACA ribonucleoprotein complex subunit 4
Alternative Name
Nucleolar protein AT band 60B
Protein minifly
Protein minifly
Location in the cell
Mitochondrial Reliability : 1.636 Nuclear Reliability : 1.86
Sequence
CDS
ATGACGGAGGTACTGCAGCCCGGTAACGAGACACTTTCTGAGAAGAAAAAGAAAAAAAACAAGGATGGTGTTTCGTTGGGAGCTTTTCAGACGATTGGCGACTTCAAAATCGAACCTTCGGAGAGCGTGAAGAAGCTGGACACCGCTTACTGGCCTTTATTGTTGAAAAACTTCGATCGCTTAAATGTGCGCACTAACCATTACACGCCGCTACCATTCGGCAACTCGCCTTTAAAACGCCCCATCTCTGATTACGTCAAATCTGGCTTCATCAACGTCGACAAACCGAGCAACCCCAGCTCCCACGAAGTAGTTTCATGGATAAAACGTATTTTAAAAGTCGAAAAGACTGGTCATTCGGGCACGCTCGATCCGAAAGTTACGGGTTGCCTTATCGTTTGCATAGACCGCGCCACGAGACTCGTCAAATCACAACAGAACGCCGGCAAAGAGTACGTAGCCGTTTTCAACTTGCATTCAGCCGTAGAGAATATTCAAAAAGTCACACAGGGCTTGGAGAAGCTGCGTGGAGCTCTATTCCAGAGGCCCCCGCTTATATCGGCCGTGAAGCGACAACTTCGTGTCAGGACGGTGTACGACAGTAAACTCCTGGATTTCGACCAGGACAAGAATATTGGGGTGTTCTGGGTTAGTTGCGAGGCTGGATCGTATATCCGGACGATGTGTGTGCATTTAGGTCTGATGCTTGGCGTGGGAGGTCAGATGATCGAACTCAGACGAGTACGGTCCGGCATTCAGGGCGAGAAGGAAGGCATGGTCACGATGCACGACATCCTAGACGCACAATGGTCATATGAAAACCACAAAGACGAGACTTACCTTCGTCGAGTAATCAAACCTCTGGAAGGCCTCCTGATTGCTCACAAAAGGATATTTATCAAAGATAGCGCCGTTAATGCTGTTTGCTACGGTGCTAAAGTGCTGTTACCAGGTATTTTAAGATACGAAGACGGCATCGAAGTTGACCAAGAAATCGTAATAGTCACCACTAAAGGTGAGGCAGTAGCGTTGGCCGTTGCTCTGATGACCACGTCTACGATGGCCTCATGCGATCATGGTGTAGCCGCGAAACTGAAGCGTGTCATAATGGAACGAGACACGTACCCCAGAAAATGGGGTCTCGGACCGAAAGCTTCACAGAAAAAAATCCTCATACAACAAGGGAAACTTGACAAATATGGCAAACCAAATGAGAACACACCAAAAGAATGGTTAAACAGTTACGTCAACTATAATGTTAAGAAAGAACCGGAGAACGGTGACGGTGTCGAGGAAGGCAGCAGAAAGCGAACGGCAAGCACAGCTAACGCGGAAGATCCCGATGTCTCCATAGAGGTGAAGCAGGAAAAGAAGAAGAAGAAGAAAAAGAGAGATAGTGAAGCCCCGTAA
Protein
MTEVLQPGNETLSEKKKKKNKDGVSLGAFQTIGDFKIEPSESVKKLDTAYWPLLLKNFDRLNVRTNHYTPLPFGNSPLKRPISDYVKSGFINVDKPSNPSSHEVVSWIKRILKVEKTGHSGTLDPKVTGCLIVCIDRATRLVKSQQNAGKEYVAVFNLHSAVENIQKVTQGLEKLRGALFQRPPLISAVKRQLRVRTVYDSKLLDFDQDKNIGVFWVSCEAGSYIRTMCVHLGLMLGVGGQMIELRRVRSGIQGEKEGMVTMHDILDAQWSYENHKDETYLRRVIKPLEGLLIAHKRIFIKDSAVNAVCYGAKVLLPGILRYEDGIEVDQEIVIVTTKGEAVALAVALMTTSTMASCDHGVAAKLKRVIMERDTYPRKWGLGPKASQKKILIQQGKLDKYGKPNENTPKEWLNSYVNYNVKKEPENGDGVEEGSRKRTASTANAEDPDVSIEVKQEKKKKKKKRDSEAP
Summary
Description
Plays a central role in ribosomal RNA processing. Probable catalytic subunit of H/ACA small nucleolar ribonucleoprotein (H/ACA snoRNP) complex, which catalyzes pseudouridylation of rRNA. This involves the isomerization of uridine such that the ribose is subsequently attached to C5, instead of the normal N1. Pseudouridine ('psi') residues may serve to stabilize the conformation of rRNAs. Required for maintenance of the germline stem cell lineage during spermatogenesis.
Catalytic Activity
a uridine in RNA = a psi-uridine in RNA
Subunit
Component of the small nucleolar ribonucleoprotein particle containing H/ACA-type snoRNAs (H/ACA snoRNPs).
Similarity
Belongs to the pseudouridine synthase TruB family.
Belongs to the type-B carboxylesterase/lipase family.
Belongs to the type-B carboxylesterase/lipase family.
Keywords
Alternative splicing
Complete proteome
Isomerase
Nucleus
Phosphoprotein
Reference proteome
Ribonucleoprotein
Ribosome biogenesis
RNA-binding
rRNA processing
Feature
chain H/ACA ribonucleoprotein complex subunit 4
splice variant In isoform C.
splice variant In isoform C.
Uniprot
H9J2U6
A0A212EMT1
A0A0N1PGC5
A0A0L7LV79
A0A2H1VJ55
A0A2A4K2B0
+ More
A0A194Q729 A0A2W1BIJ1 A0A182J790 U5EZN4 A0A182MIS3 A0A182R2S0 A0A182WAR9 A0A182SM12 A0A084WIA0 A0A182WZW6 A0A182QMF4 A0A182VIW2 A0A182JVE8 Q7PMU4 A0A182P609 A0A182NNA7 B0W1L7 B0WI05 A0A1S4F520 W8CCM1 Q0IG45 A0A2M4A8W0 A0A182FTX0 A0A2M4A8U3 A0A2M4A8S4 A0A2M3Z206 A0A2M4BK78 A0A2M4BK29 A0A2M4BKR6 A0A2M3ZGF4 W5JHW9 A0A034VRL9 T1DQ85 A0A1Q3EWE0 A0A1Q3EWB2 A0A182GN81 A0A0A1WIC1 B4PAW0 B3NQG5 A0A023EWJ2 A0A0K8UWZ2 H5V894 B4QBJ6 C0P8M5 E1JGV6 O44081 B4I2E9 A0A1L8DSB9 A0A1W4UMH6 A0A1J1IQ89 B4J4S2 A0A3B0IYW7 A0A1I8PCY8 A0A1A9UES3 A0A1B0CIC5 A0A0L0C603 T1PDS7 A0A1A9Y4U5 D2CFW9 B3MDN0 A0A336LNS5 D3TP71 A0A1I8MBT6 A0A0Q9WPI8 B4H6A4 A0A336LIQ0 A0A1Y1LPL5 B4KM31 B4LK39 A0A0M4EVY8 A0A1A9ZQU3 B5DZ83 A0A1B0G7F7 N6T638 U4U617 E9HCX8 A0A0P4WPK2 A0A0P5PGT8 A0A0P5GBW8 A0A0P5KSS2 A0A0P5FKC1 A0A0P5ED15 A0A0P5B0W9 A0A0P5ZNN2 A0A0P4WPN5 A0A0P6I206 A0A0P5FML6 A0A0P5FTI3 A0A0P6FCW9 A0A0P5K1J9 A0A0P5W8B6 A0A0P4ZXP1 A0A0P5A4Y4 A0A0P5MHG3
A0A194Q729 A0A2W1BIJ1 A0A182J790 U5EZN4 A0A182MIS3 A0A182R2S0 A0A182WAR9 A0A182SM12 A0A084WIA0 A0A182WZW6 A0A182QMF4 A0A182VIW2 A0A182JVE8 Q7PMU4 A0A182P609 A0A182NNA7 B0W1L7 B0WI05 A0A1S4F520 W8CCM1 Q0IG45 A0A2M4A8W0 A0A182FTX0 A0A2M4A8U3 A0A2M4A8S4 A0A2M3Z206 A0A2M4BK78 A0A2M4BK29 A0A2M4BKR6 A0A2M3ZGF4 W5JHW9 A0A034VRL9 T1DQ85 A0A1Q3EWE0 A0A1Q3EWB2 A0A182GN81 A0A0A1WIC1 B4PAW0 B3NQG5 A0A023EWJ2 A0A0K8UWZ2 H5V894 B4QBJ6 C0P8M5 E1JGV6 O44081 B4I2E9 A0A1L8DSB9 A0A1W4UMH6 A0A1J1IQ89 B4J4S2 A0A3B0IYW7 A0A1I8PCY8 A0A1A9UES3 A0A1B0CIC5 A0A0L0C603 T1PDS7 A0A1A9Y4U5 D2CFW9 B3MDN0 A0A336LNS5 D3TP71 A0A1I8MBT6 A0A0Q9WPI8 B4H6A4 A0A336LIQ0 A0A1Y1LPL5 B4KM31 B4LK39 A0A0M4EVY8 A0A1A9ZQU3 B5DZ83 A0A1B0G7F7 N6T638 U4U617 E9HCX8 A0A0P4WPK2 A0A0P5PGT8 A0A0P5GBW8 A0A0P5KSS2 A0A0P5FKC1 A0A0P5ED15 A0A0P5B0W9 A0A0P5ZNN2 A0A0P4WPN5 A0A0P6I206 A0A0P5FML6 A0A0P5FTI3 A0A0P6FCW9 A0A0P5K1J9 A0A0P5W8B6 A0A0P4ZXP1 A0A0P5A4Y4 A0A0P5MHG3
EC Number
5.4.99.-
Pubmed
19121390
22118469
26354079
26227816
28756777
24438588
+ More
12364791 14747013 17210077 24495485 17510324 20920257 23761445 25348373 26483478 25830018 17994087 17550304 24945155 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 9829824 10087258 17328797 12537569 12645924 18327897 26108605 18362917 19820115 20353571 25315136 28004739 15632085 23537049 21292972
12364791 14747013 17210077 24495485 17510324 20920257 23761445 25348373 26483478 25830018 17994087 17550304 24945155 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 9829824 10087258 17328797 12537569 12645924 18327897 26108605 18362917 19820115 20353571 25315136 28004739 15632085 23537049 21292972
EMBL
BABH01036910
AGBW02013797
OWR42805.1
KQ460735
KPJ12619.1
JTDY01000027
+ More
KOB79335.1 ODYU01002860 SOQ40865.1 NWSH01000242 PCG78068.1 KQ459580 KPI99200.1 KZ150307 PZC71493.1 GANO01000464 JAB59407.1 AXCM01000074 ATLV01023923 KE525347 KFB49944.1 AXCN02001904 AAAB01008968 EAA13199.5 DS231822 EDS25815.1 DS231941 EDS28087.1 GAMC01003457 GAMC01003456 JAC03100.1 CH477274 EAT45166.1 GGFK01003888 MBW37209.1 GGFK01003868 MBW37189.1 GGFK01003883 MBW37204.1 GGFM01001799 MBW22550.1 GGFJ01004242 MBW53383.1 GGFJ01004241 MBW53382.1 GGFJ01004247 MBW53388.1 GGFM01006820 MBW27571.1 ADMH02001457 ETN62500.1 GAKP01012995 GAKP01012991 JAC45961.1 GAMD01001305 JAB00286.1 GFDL01015459 JAV19586.1 GFDL01015455 JAV19590.1 JXUM01014594 JXUM01014595 KQ560407 KXJ82572.1 GBXI01015886 JAC98405.1 CM000158 EDW92500.1 CH954179 EDV56968.1 GAPW01000719 JAC12879.1 GDHF01021454 JAI30860.1 BT133195 AFA28436.1 CM000362 CM002911 EDX08514.1 KMY96287.1 BT064644 ACN29341.1 AE013599 ACZ94558.1 ACZ94561.1 AF017230 AF089837 AF097634 DQ841550 DQ857345 DQ857346 AY089408 BT030987 CH480820 EDW53944.1 GFDF01004869 JAV09215.1 CVRI01000057 CRL02405.1 CH916367 EDW00618.1 OUUW01000001 SPP73404.1 AJWK01013184 AJWK01013185 JRES01000857 KNC27710.1 KA646849 AFP61478.1 DS497670 EFA13038.1 CH902619 EDV37493.1 UFQT01000092 SSX19580.1 EZ423223 ADD19499.1 CH963850 KRF98116.1 CH479213 EDW33328.1 UFQS01002236 UFQT01002236 SSX13562.1 SSX32989.1 GEZM01050460 JAV75583.1 CH933808 EDW10820.1 CH940648 EDW60628.1 CP012524 ALC41870.1 CM000071 EDY69554.2 CCAG010019065 APGK01050961 APGK01050962 KB741180 ENN73118.1 KB631687 ERL85380.1 GL732621 EFX70431.1 GDIP01253467 JAI69934.1 GDIQ01128641 JAL23085.1 GDIQ01250590 JAK01135.1 GDIQ01181258 JAK70467.1 GDIQ01253294 JAJ98430.1 GDIP01144646 JAJ78756.1 GDIP01191595 JAJ31807.1 GDIP01041404 JAM62311.1 GDIP01253421 JAI69980.1 GDIQ01018381 JAN76356.1 GDIQ01253295 JAJ98429.1 GDIQ01250591 JAK01134.1 GDIQ01050666 JAN44071.1 GDIQ01196047 JAK55678.1 GDIP01089809 JAM13906.1 GDIP01210975 JAJ12427.1 GDIP01207534 JAJ15868.1 GDIQ01159173 JAK92552.1
KOB79335.1 ODYU01002860 SOQ40865.1 NWSH01000242 PCG78068.1 KQ459580 KPI99200.1 KZ150307 PZC71493.1 GANO01000464 JAB59407.1 AXCM01000074 ATLV01023923 KE525347 KFB49944.1 AXCN02001904 AAAB01008968 EAA13199.5 DS231822 EDS25815.1 DS231941 EDS28087.1 GAMC01003457 GAMC01003456 JAC03100.1 CH477274 EAT45166.1 GGFK01003888 MBW37209.1 GGFK01003868 MBW37189.1 GGFK01003883 MBW37204.1 GGFM01001799 MBW22550.1 GGFJ01004242 MBW53383.1 GGFJ01004241 MBW53382.1 GGFJ01004247 MBW53388.1 GGFM01006820 MBW27571.1 ADMH02001457 ETN62500.1 GAKP01012995 GAKP01012991 JAC45961.1 GAMD01001305 JAB00286.1 GFDL01015459 JAV19586.1 GFDL01015455 JAV19590.1 JXUM01014594 JXUM01014595 KQ560407 KXJ82572.1 GBXI01015886 JAC98405.1 CM000158 EDW92500.1 CH954179 EDV56968.1 GAPW01000719 JAC12879.1 GDHF01021454 JAI30860.1 BT133195 AFA28436.1 CM000362 CM002911 EDX08514.1 KMY96287.1 BT064644 ACN29341.1 AE013599 ACZ94558.1 ACZ94561.1 AF017230 AF089837 AF097634 DQ841550 DQ857345 DQ857346 AY089408 BT030987 CH480820 EDW53944.1 GFDF01004869 JAV09215.1 CVRI01000057 CRL02405.1 CH916367 EDW00618.1 OUUW01000001 SPP73404.1 AJWK01013184 AJWK01013185 JRES01000857 KNC27710.1 KA646849 AFP61478.1 DS497670 EFA13038.1 CH902619 EDV37493.1 UFQT01000092 SSX19580.1 EZ423223 ADD19499.1 CH963850 KRF98116.1 CH479213 EDW33328.1 UFQS01002236 UFQT01002236 SSX13562.1 SSX32989.1 GEZM01050460 JAV75583.1 CH933808 EDW10820.1 CH940648 EDW60628.1 CP012524 ALC41870.1 CM000071 EDY69554.2 CCAG010019065 APGK01050961 APGK01050962 KB741180 ENN73118.1 KB631687 ERL85380.1 GL732621 EFX70431.1 GDIP01253467 JAI69934.1 GDIQ01128641 JAL23085.1 GDIQ01250590 JAK01135.1 GDIQ01181258 JAK70467.1 GDIQ01253294 JAJ98430.1 GDIP01144646 JAJ78756.1 GDIP01191595 JAJ31807.1 GDIP01041404 JAM62311.1 GDIP01253421 JAI69980.1 GDIQ01018381 JAN76356.1 GDIQ01253295 JAJ98429.1 GDIQ01250591 JAK01134.1 GDIQ01050666 JAN44071.1 GDIQ01196047 JAK55678.1 GDIP01089809 JAM13906.1 GDIP01210975 JAJ12427.1 GDIP01207534 JAJ15868.1 GDIQ01159173 JAK92552.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000037510
UP000218220
UP000053268
+ More
UP000075880 UP000075883 UP000075900 UP000075920 UP000075901 UP000030765 UP000076407 UP000075886 UP000075903 UP000075881 UP000007062 UP000075885 UP000075884 UP000002320 UP000008820 UP000069272 UP000000673 UP000069940 UP000249989 UP000002282 UP000008711 UP000000304 UP000000803 UP000001292 UP000192221 UP000183832 UP000001070 UP000268350 UP000095300 UP000078200 UP000092461 UP000037069 UP000092443 UP000007266 UP000007801 UP000095301 UP000007798 UP000008744 UP000009192 UP000008792 UP000092553 UP000092445 UP000001819 UP000092444 UP000019118 UP000030742 UP000000305
UP000075880 UP000075883 UP000075900 UP000075920 UP000075901 UP000030765 UP000076407 UP000075886 UP000075903 UP000075881 UP000007062 UP000075885 UP000075884 UP000002320 UP000008820 UP000069272 UP000000673 UP000069940 UP000249989 UP000002282 UP000008711 UP000000304 UP000000803 UP000001292 UP000192221 UP000183832 UP000001070 UP000268350 UP000095300 UP000078200 UP000092461 UP000037069 UP000092443 UP000007266 UP000007801 UP000095301 UP000007798 UP000008744 UP000009192 UP000008792 UP000092553 UP000092445 UP000001819 UP000092444 UP000019118 UP000030742 UP000000305
Interpro
IPR004521
Uncharacterised_CHP00451
+ More
IPR032819 TruB_C
IPR002501 PsdUridine_synth_N
IPR002478 PUA
IPR004802 tRNA_PsdUridine_synth_B_fam
IPR015947 PUA-like_sf
IPR020103 PsdUridine_synth_cat_dom_sf
IPR012960 Dyskerin-like
IPR019819 Carboxylesterase_B_CS
IPR029058 AB_hydrolase
IPR002018 CarbesteraseB
IPR019826 Carboxylesterase_B_AS
IPR032819 TruB_C
IPR002501 PsdUridine_synth_N
IPR002478 PUA
IPR004802 tRNA_PsdUridine_synth_B_fam
IPR015947 PUA-like_sf
IPR020103 PsdUridine_synth_cat_dom_sf
IPR012960 Dyskerin-like
IPR019819 Carboxylesterase_B_CS
IPR029058 AB_hydrolase
IPR002018 CarbesteraseB
IPR019826 Carboxylesterase_B_AS
Gene 3D
ProteinModelPortal
H9J2U6
A0A212EMT1
A0A0N1PGC5
A0A0L7LV79
A0A2H1VJ55
A0A2A4K2B0
+ More
A0A194Q729 A0A2W1BIJ1 A0A182J790 U5EZN4 A0A182MIS3 A0A182R2S0 A0A182WAR9 A0A182SM12 A0A084WIA0 A0A182WZW6 A0A182QMF4 A0A182VIW2 A0A182JVE8 Q7PMU4 A0A182P609 A0A182NNA7 B0W1L7 B0WI05 A0A1S4F520 W8CCM1 Q0IG45 A0A2M4A8W0 A0A182FTX0 A0A2M4A8U3 A0A2M4A8S4 A0A2M3Z206 A0A2M4BK78 A0A2M4BK29 A0A2M4BKR6 A0A2M3ZGF4 W5JHW9 A0A034VRL9 T1DQ85 A0A1Q3EWE0 A0A1Q3EWB2 A0A182GN81 A0A0A1WIC1 B4PAW0 B3NQG5 A0A023EWJ2 A0A0K8UWZ2 H5V894 B4QBJ6 C0P8M5 E1JGV6 O44081 B4I2E9 A0A1L8DSB9 A0A1W4UMH6 A0A1J1IQ89 B4J4S2 A0A3B0IYW7 A0A1I8PCY8 A0A1A9UES3 A0A1B0CIC5 A0A0L0C603 T1PDS7 A0A1A9Y4U5 D2CFW9 B3MDN0 A0A336LNS5 D3TP71 A0A1I8MBT6 A0A0Q9WPI8 B4H6A4 A0A336LIQ0 A0A1Y1LPL5 B4KM31 B4LK39 A0A0M4EVY8 A0A1A9ZQU3 B5DZ83 A0A1B0G7F7 N6T638 U4U617 E9HCX8 A0A0P4WPK2 A0A0P5PGT8 A0A0P5GBW8 A0A0P5KSS2 A0A0P5FKC1 A0A0P5ED15 A0A0P5B0W9 A0A0P5ZNN2 A0A0P4WPN5 A0A0P6I206 A0A0P5FML6 A0A0P5FTI3 A0A0P6FCW9 A0A0P5K1J9 A0A0P5W8B6 A0A0P4ZXP1 A0A0P5A4Y4 A0A0P5MHG3
A0A194Q729 A0A2W1BIJ1 A0A182J790 U5EZN4 A0A182MIS3 A0A182R2S0 A0A182WAR9 A0A182SM12 A0A084WIA0 A0A182WZW6 A0A182QMF4 A0A182VIW2 A0A182JVE8 Q7PMU4 A0A182P609 A0A182NNA7 B0W1L7 B0WI05 A0A1S4F520 W8CCM1 Q0IG45 A0A2M4A8W0 A0A182FTX0 A0A2M4A8U3 A0A2M4A8S4 A0A2M3Z206 A0A2M4BK78 A0A2M4BK29 A0A2M4BKR6 A0A2M3ZGF4 W5JHW9 A0A034VRL9 T1DQ85 A0A1Q3EWE0 A0A1Q3EWB2 A0A182GN81 A0A0A1WIC1 B4PAW0 B3NQG5 A0A023EWJ2 A0A0K8UWZ2 H5V894 B4QBJ6 C0P8M5 E1JGV6 O44081 B4I2E9 A0A1L8DSB9 A0A1W4UMH6 A0A1J1IQ89 B4J4S2 A0A3B0IYW7 A0A1I8PCY8 A0A1A9UES3 A0A1B0CIC5 A0A0L0C603 T1PDS7 A0A1A9Y4U5 D2CFW9 B3MDN0 A0A336LNS5 D3TP71 A0A1I8MBT6 A0A0Q9WPI8 B4H6A4 A0A336LIQ0 A0A1Y1LPL5 B4KM31 B4LK39 A0A0M4EVY8 A0A1A9ZQU3 B5DZ83 A0A1B0G7F7 N6T638 U4U617 E9HCX8 A0A0P4WPK2 A0A0P5PGT8 A0A0P5GBW8 A0A0P5KSS2 A0A0P5FKC1 A0A0P5ED15 A0A0P5B0W9 A0A0P5ZNN2 A0A0P4WPN5 A0A0P6I206 A0A0P5FML6 A0A0P5FTI3 A0A0P6FCW9 A0A0P5K1J9 A0A0P5W8B6 A0A0P4ZXP1 A0A0P5A4Y4 A0A0P5MHG3
PDB
3UAI
E-value=2.01393e-161,
Score=1461
Ontologies
GO
PANTHER
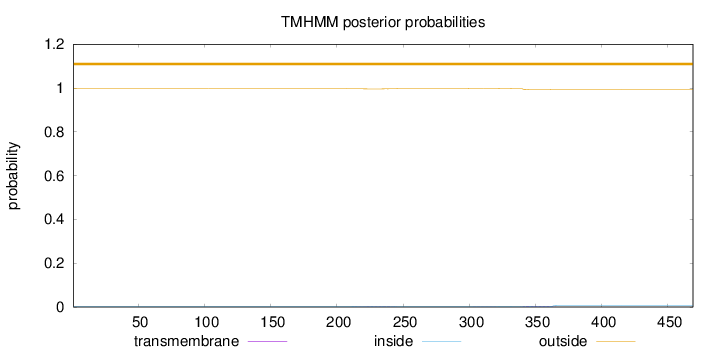
Topology
Subcellular location
Length:
469
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.13896
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00329
outside
1 - 469
Population Genetic Test Statistics
Pi
330.514046
Theta
210.769896
Tajima's D
1.439667
CLR
0
CSRT
0.774461276936153
Interpretation
Uncertain
