Gene
KWMTBOMO14750
Pre Gene Modal
BGIBMGA003821
Annotation
PREDICTED:_single-minded_homolog_1_[Bombyx_mori]
Full name
Protein single-minded
Transcription factor
Location in the cell
Mitochondrial Reliability : 2.361
Sequence
CDS
ATGGCCGACGCAGGCCGCAAGGAGCTGCGGCAGCTGTCGCAGGTCGCGGCAAACATCATGAAGGAGAAAAGCAAAAACGCGGCACGATCCAGGAGGGAAAAAGAGAACACAGAATTCTTGGAGCTGGCGAAGCTGCTGCCTCTGCCTTCTGCGATCACCTCGCAGCTGGACAAGGCCTCCGTTATAAGACTAACGACCAGCTACCTGAAGATGCGCCAGGTTTTTCCTGATGGCTTGGGGGACGCGTGGGGAGCGGCGCCACCACCACCGCAACCCCGCGAATTGTCGATACGAGAGCTCGGGTCGCATTTGCTCCAAACTTTGGACGGTTTTATATTCGTAGTGGCTCCCGATGGCAAAATTATGTACATCAGCGAAACTGCGTCAGTACATCTTGGCTTGAGTCAGGTGGAACTGACCGGCAACTCTATATACGAGTACATCCACCAAGCAGACCACGAGGAGATGAATGCAGTACTGAGTCTCCACCACCCTCATTCATACCACGGCCACCAGTATCCTTCAGTTGGGTATCCGGTGGGAGGGACATGGGGTCCGACAGTCGATGCGGAGTGTGAAAGAGCGTTCTTCATCAGAATGAAATGTGTGCTGGCGAAGAGGAACGCCGGCTTAACCACCTCAGGATACAAGGTGATCCACTGCTCCGGCTACCTAAGAGTTCGGCGGTTTGGTGAGGGCTCCGCCCCCCTCGGGCTGGTCGCGGTGGGTCACTCCCTCCCGCCCTCCGCCGTCACAGAACTCAAGCTGCACTCCAACATGTTCATGTTCCGAGCGTCGTTGGACATGCGGCTGATCTTCCTGGATGCAAGACTTCGTAGTGCCTCAATCGATAAATAA
Protein
MADAGRKELRQLSQVAANIMKEKSKNAARSRREKENTEFLELAKLLPLPSAITSQLDKASVIRLTTSYLKMRQVFPDGLGDAWGAAPPPPQPRELSIRELGSHLLQTLDGFIFVVAPDGKIMYISETASVHLGLSQVELTGNSIYEYIHQADHEEMNAVLSLHHPHSYHGHQYPSVGYPVGGTWGPTVDAECERAFFIRMKCVLAKRNAGLTTSGYKVIHCSGYLRVRRFGEGSAPLGLVAVGHSLPPSAVTELKLHSNMFMFRASLDMRLIFLDARLRSASIDK
Summary
Description
Transcription factor that functions as a master developmental regulator controlling midline development of the ventral nerve cord. Required to correctly specify the formation of the central brain complex, which controls walking behavior. Also required for correct patterning of the embryonic genital disk and anal pad anlage. Plays a role in synapse development.
Subunit
Efficient DNA binding requires dimerization with another bHLH protein.
Miscellaneous
Mutations result in the loss of the precursor cells that give rise to midline cells of the embryonic central nervous system.
Keywords
Alternative splicing
Complete proteome
Developmental protein
Differentiation
DNA-binding
Neurogenesis
Nucleus
Polymorphism
Reference proteome
Repeat
Transcription
Transcription regulation
Feature
chain Protein single-minded
splice variant In isoform B.
splice variant In isoform B.
Uniprot
A0A194Q1J7
A0A3S2NEK8
A0A2H1VVZ3
A0A2A4K220
A0A212F8X1
A0A0N1IFA4
+ More
H9J2T4 A0A2J7QQL4 A0A2J7QQN0 A0A067QLY0 A0A2P8ZFK6 A0A1Y1JWW2 A0A2R7WII5 A0A023F2C6 A0A0F7II89 A0A146MAW1 A0A146KRX5 A0A0P5V8R1 A0A1B6K0I5 A0A1B6IIL4 T1HC33 A0A1B6LAG7 A0A0P6BZK7 A0A0P6CLA7 A0A0B6VJJ6 E9FY44 T1IKN0 E0VC62 A0A3L8SZW5 A0A226PUL3 Q25C45 A0A2U3X830 A0A3L7I4L7 A0A1B6CUM8 V3ZDU5 L5JM98 A0A1B6CA85 A0A060XJA4 H3B852 B4NG65 B4G5M2 A0A0M5J5E5 B5DXT9 A0A1S3JVS7 A0A3Q7XWA1 C3XTH0 B3LX41 A0A3B0KMU7 A0A067Y793 A0A0L0C053 B4M6L5 A0A0B4KH05 A0A1S3JVT4 P05709-2 P05709 A0A3B0KRN7 A0A3B0KVA8 B3P185 A0A1S3JV10 A0A0B4KFY1 B4R1W3 A0A1A9V500 A0A1A9W571 B4K8H0 B4JEV3 Q1PHQ4 Q5R8E1 A0A2C9KA40 A0A1W4UKT6 A0A1B0FK96 A0A1A9X5X0 A0A1B0AWL8 A0A0B6ZMK6 A0A3B3W0U5 A0A3B0KHJ5 B4PPW7 A0A0R1E2U3 G1KJZ0 F7GDY6 A0A1I8NLQ3 A0A0Q3MJN7 A0A2S2P376 A0A1L8G9C3 A0A3B4ZHA9 A0A0R1E2V5 A0A1B0AKC2 A0A0R1E3F6 A0A0R1E6T5 A0A338P771 A0A3B5KJW1 F6U9K2 A0A0R1E2S5 R0LKM3 A0A151NTB9 A0A3Q1F0W8 A0A1U7RM06 A0A0R1E495 A0A226E3G3 A0A091GA89 H0ZPV8
H9J2T4 A0A2J7QQL4 A0A2J7QQN0 A0A067QLY0 A0A2P8ZFK6 A0A1Y1JWW2 A0A2R7WII5 A0A023F2C6 A0A0F7II89 A0A146MAW1 A0A146KRX5 A0A0P5V8R1 A0A1B6K0I5 A0A1B6IIL4 T1HC33 A0A1B6LAG7 A0A0P6BZK7 A0A0P6CLA7 A0A0B6VJJ6 E9FY44 T1IKN0 E0VC62 A0A3L8SZW5 A0A226PUL3 Q25C45 A0A2U3X830 A0A3L7I4L7 A0A1B6CUM8 V3ZDU5 L5JM98 A0A1B6CA85 A0A060XJA4 H3B852 B4NG65 B4G5M2 A0A0M5J5E5 B5DXT9 A0A1S3JVS7 A0A3Q7XWA1 C3XTH0 B3LX41 A0A3B0KMU7 A0A067Y793 A0A0L0C053 B4M6L5 A0A0B4KH05 A0A1S3JVT4 P05709-2 P05709 A0A3B0KRN7 A0A3B0KVA8 B3P185 A0A1S3JV10 A0A0B4KFY1 B4R1W3 A0A1A9V500 A0A1A9W571 B4K8H0 B4JEV3 Q1PHQ4 Q5R8E1 A0A2C9KA40 A0A1W4UKT6 A0A1B0FK96 A0A1A9X5X0 A0A1B0AWL8 A0A0B6ZMK6 A0A3B3W0U5 A0A3B0KHJ5 B4PPW7 A0A0R1E2U3 G1KJZ0 F7GDY6 A0A1I8NLQ3 A0A0Q3MJN7 A0A2S2P376 A0A1L8G9C3 A0A3B4ZHA9 A0A0R1E2V5 A0A1B0AKC2 A0A0R1E3F6 A0A0R1E6T5 A0A338P771 A0A3B5KJW1 F6U9K2 A0A0R1E2S5 R0LKM3 A0A151NTB9 A0A3Q1F0W8 A0A1U7RM06 A0A0R1E495 A0A226E3G3 A0A091GA89 H0ZPV8
Pubmed
26354079
22118469
19121390
24845553
29403074
28004739
+ More
25474469 26823975 25447925 21292972 20566863 30282656 24621616 16720876 29704459 23254933 23258410 24755649 9215903 17994087 15632085 18563158 24959341 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 9840810 12221007 12537569 1760843 3345560 23644463 16933975 15562597 17550304 18464734 27762356 19468303 21551351 20431018 22293439 20360741
25474469 26823975 25447925 21292972 20566863 30282656 24621616 16720876 29704459 23254933 23258410 24755649 9215903 17994087 15632085 18563158 24959341 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 9840810 12221007 12537569 1760843 3345560 23644463 16933975 15562597 17550304 18464734 27762356 19468303 21551351 20431018 22293439 20360741
EMBL
KQ459580
KPI99198.1
RSAL01000072
RVE49077.1
ODYU01004781
SOQ45021.1
+ More
NWSH01000242 PCG78066.1 AGBW02009662 OWR50177.1 KQ460735 KPJ12617.1 BABH01036902 BABH01036903 BABH01036904 NEVH01012087 PNF30878.1 PNF30877.1 KK853270 KDR09145.1 PYGN01000072 PSN55283.1 GEZM01098957 JAV53613.1 KK854838 PTY19151.1 GBBI01002945 JAC15767.1 KP147936 AKG92773.1 GDHC01002447 JAQ16182.1 GDHC01020642 JAP97986.1 GDIP01105491 JAL98223.1 GECU01002752 JAT04955.1 GECU01020951 JAS86755.1 ACPB03006262 GEBQ01019265 JAT20712.1 GDIP01181764 GDIP01007128 JAM96587.1 GDIQ01090729 JAN04008.1 AB981593 BAQ21977.1 GL732527 EFX87839.1 JH430577 DS235047 EEB10968.1 QUSF01000002 RLW12415.1 AWGT02000016 OXB83170.1 AB236150 BAE87099.1 RAZU01000101 RLQ72890.1 GEDC01020263 JAS17035.1 KB202619 ESO89288.1 KB031158 ELK00052.1 GEDC01026937 JAS10361.1 FR905472 CDQ79551.1 AFYH01063801 AFYH01063802 CH964251 EDW83282.2 CH479179 EDW24888.1 CP012526 ALC47531.1 CM000070 EDY67905.2 GG666463 EEN68596.1 CH902617 EDV41641.2 OUUW01000013 SPP87909.1 KC305631 AGX25236.1 JRES01001168 KNC24839.1 CH940652 EDW59291.2 AE014297 AGB95909.1 AF071934 AY129457 M19020 SPP87911.1 SPP87908.1 CH954181 EDV49274.1 AGB95910.1 CM000364 EDX13115.1 CH933806 EDW14369.2 CH916369 EDV93234.1 DQ431043 ABD97275.1 CR859812 CAH91969.1 CCAG010014365 JXJN01004815 HACG01022898 CEK69763.1 SPP87910.1 CM000160 EDW97194.1 KRK03579.1 LMAW01001780 KQK82664.1 GGMR01011183 MBY23802.1 CM004474 OCT80433.1 KRK03580.1 KRK03581.1 KRK03583.1 KRK03585.1 AC165961 CU207273 AAMC01009000 AAMC01009001 KRK03584.1 KB742627 EOB06249.1 AKHW03002098 KYO40028.1 KRK03582.1 LNIX01000008 OXA51036.1 KL447928 KFO78838.1 ABQF01000555
NWSH01000242 PCG78066.1 AGBW02009662 OWR50177.1 KQ460735 KPJ12617.1 BABH01036902 BABH01036903 BABH01036904 NEVH01012087 PNF30878.1 PNF30877.1 KK853270 KDR09145.1 PYGN01000072 PSN55283.1 GEZM01098957 JAV53613.1 KK854838 PTY19151.1 GBBI01002945 JAC15767.1 KP147936 AKG92773.1 GDHC01002447 JAQ16182.1 GDHC01020642 JAP97986.1 GDIP01105491 JAL98223.1 GECU01002752 JAT04955.1 GECU01020951 JAS86755.1 ACPB03006262 GEBQ01019265 JAT20712.1 GDIP01181764 GDIP01007128 JAM96587.1 GDIQ01090729 JAN04008.1 AB981593 BAQ21977.1 GL732527 EFX87839.1 JH430577 DS235047 EEB10968.1 QUSF01000002 RLW12415.1 AWGT02000016 OXB83170.1 AB236150 BAE87099.1 RAZU01000101 RLQ72890.1 GEDC01020263 JAS17035.1 KB202619 ESO89288.1 KB031158 ELK00052.1 GEDC01026937 JAS10361.1 FR905472 CDQ79551.1 AFYH01063801 AFYH01063802 CH964251 EDW83282.2 CH479179 EDW24888.1 CP012526 ALC47531.1 CM000070 EDY67905.2 GG666463 EEN68596.1 CH902617 EDV41641.2 OUUW01000013 SPP87909.1 KC305631 AGX25236.1 JRES01001168 KNC24839.1 CH940652 EDW59291.2 AE014297 AGB95909.1 AF071934 AY129457 M19020 SPP87911.1 SPP87908.1 CH954181 EDV49274.1 AGB95910.1 CM000364 EDX13115.1 CH933806 EDW14369.2 CH916369 EDV93234.1 DQ431043 ABD97275.1 CR859812 CAH91969.1 CCAG010014365 JXJN01004815 HACG01022898 CEK69763.1 SPP87910.1 CM000160 EDW97194.1 KRK03579.1 LMAW01001780 KQK82664.1 GGMR01011183 MBY23802.1 CM004474 OCT80433.1 KRK03580.1 KRK03581.1 KRK03583.1 KRK03585.1 AC165961 CU207273 AAMC01009000 AAMC01009001 KRK03584.1 KB742627 EOB06249.1 AKHW03002098 KYO40028.1 KRK03582.1 LNIX01000008 OXA51036.1 KL447928 KFO78838.1 ABQF01000555
Proteomes
UP000053268
UP000283053
UP000218220
UP000007151
UP000053240
UP000005204
+ More
UP000235965 UP000027135 UP000245037 UP000015103 UP000000305 UP000009046 UP000276834 UP000198419 UP000245341 UP000273346 UP000030746 UP000010552 UP000193380 UP000008672 UP000007798 UP000008744 UP000092553 UP000001819 UP000085678 UP000286642 UP000001554 UP000007801 UP000268350 UP000037069 UP000008792 UP000000803 UP000008711 UP000000304 UP000078200 UP000091820 UP000009192 UP000001070 UP000076420 UP000192221 UP000092444 UP000092443 UP000092460 UP000261500 UP000002282 UP000001646 UP000002279 UP000095300 UP000051836 UP000186698 UP000261400 UP000092445 UP000000589 UP000005226 UP000008143 UP000050525 UP000257200 UP000189705 UP000198287 UP000053760 UP000007754
UP000235965 UP000027135 UP000245037 UP000015103 UP000000305 UP000009046 UP000276834 UP000198419 UP000245341 UP000273346 UP000030746 UP000010552 UP000193380 UP000008672 UP000007798 UP000008744 UP000092553 UP000001819 UP000085678 UP000286642 UP000001554 UP000007801 UP000268350 UP000037069 UP000008792 UP000000803 UP000008711 UP000000304 UP000078200 UP000091820 UP000009192 UP000001070 UP000076420 UP000192221 UP000092444 UP000092443 UP000092460 UP000261500 UP000002282 UP000001646 UP000002279 UP000095300 UP000051836 UP000186698 UP000261400 UP000092445 UP000000589 UP000005226 UP000008143 UP000050525 UP000257200 UP000189705 UP000198287 UP000053760 UP000007754
Interpro
Gene 3D
ProteinModelPortal
A0A194Q1J7
A0A3S2NEK8
A0A2H1VVZ3
A0A2A4K220
A0A212F8X1
A0A0N1IFA4
+ More
H9J2T4 A0A2J7QQL4 A0A2J7QQN0 A0A067QLY0 A0A2P8ZFK6 A0A1Y1JWW2 A0A2R7WII5 A0A023F2C6 A0A0F7II89 A0A146MAW1 A0A146KRX5 A0A0P5V8R1 A0A1B6K0I5 A0A1B6IIL4 T1HC33 A0A1B6LAG7 A0A0P6BZK7 A0A0P6CLA7 A0A0B6VJJ6 E9FY44 T1IKN0 E0VC62 A0A3L8SZW5 A0A226PUL3 Q25C45 A0A2U3X830 A0A3L7I4L7 A0A1B6CUM8 V3ZDU5 L5JM98 A0A1B6CA85 A0A060XJA4 H3B852 B4NG65 B4G5M2 A0A0M5J5E5 B5DXT9 A0A1S3JVS7 A0A3Q7XWA1 C3XTH0 B3LX41 A0A3B0KMU7 A0A067Y793 A0A0L0C053 B4M6L5 A0A0B4KH05 A0A1S3JVT4 P05709-2 P05709 A0A3B0KRN7 A0A3B0KVA8 B3P185 A0A1S3JV10 A0A0B4KFY1 B4R1W3 A0A1A9V500 A0A1A9W571 B4K8H0 B4JEV3 Q1PHQ4 Q5R8E1 A0A2C9KA40 A0A1W4UKT6 A0A1B0FK96 A0A1A9X5X0 A0A1B0AWL8 A0A0B6ZMK6 A0A3B3W0U5 A0A3B0KHJ5 B4PPW7 A0A0R1E2U3 G1KJZ0 F7GDY6 A0A1I8NLQ3 A0A0Q3MJN7 A0A2S2P376 A0A1L8G9C3 A0A3B4ZHA9 A0A0R1E2V5 A0A1B0AKC2 A0A0R1E3F6 A0A0R1E6T5 A0A338P771 A0A3B5KJW1 F6U9K2 A0A0R1E2S5 R0LKM3 A0A151NTB9 A0A3Q1F0W8 A0A1U7RM06 A0A0R1E495 A0A226E3G3 A0A091GA89 H0ZPV8
H9J2T4 A0A2J7QQL4 A0A2J7QQN0 A0A067QLY0 A0A2P8ZFK6 A0A1Y1JWW2 A0A2R7WII5 A0A023F2C6 A0A0F7II89 A0A146MAW1 A0A146KRX5 A0A0P5V8R1 A0A1B6K0I5 A0A1B6IIL4 T1HC33 A0A1B6LAG7 A0A0P6BZK7 A0A0P6CLA7 A0A0B6VJJ6 E9FY44 T1IKN0 E0VC62 A0A3L8SZW5 A0A226PUL3 Q25C45 A0A2U3X830 A0A3L7I4L7 A0A1B6CUM8 V3ZDU5 L5JM98 A0A1B6CA85 A0A060XJA4 H3B852 B4NG65 B4G5M2 A0A0M5J5E5 B5DXT9 A0A1S3JVS7 A0A3Q7XWA1 C3XTH0 B3LX41 A0A3B0KMU7 A0A067Y793 A0A0L0C053 B4M6L5 A0A0B4KH05 A0A1S3JVT4 P05709-2 P05709 A0A3B0KRN7 A0A3B0KVA8 B3P185 A0A1S3JV10 A0A0B4KFY1 B4R1W3 A0A1A9V500 A0A1A9W571 B4K8H0 B4JEV3 Q1PHQ4 Q5R8E1 A0A2C9KA40 A0A1W4UKT6 A0A1B0FK96 A0A1A9X5X0 A0A1B0AWL8 A0A0B6ZMK6 A0A3B3W0U5 A0A3B0KHJ5 B4PPW7 A0A0R1E2U3 G1KJZ0 F7GDY6 A0A1I8NLQ3 A0A0Q3MJN7 A0A2S2P376 A0A1L8G9C3 A0A3B4ZHA9 A0A0R1E2V5 A0A1B0AKC2 A0A0R1E3F6 A0A0R1E6T5 A0A338P771 A0A3B5KJW1 F6U9K2 A0A0R1E2S5 R0LKM3 A0A151NTB9 A0A3Q1F0W8 A0A1U7RM06 A0A0R1E495 A0A226E3G3 A0A091GA89 H0ZPV8
PDB
5SY7
E-value=6.02641e-59,
Score=575
Ontologies
GO
GO:0005667
GO:0005737
GO:0003700
GO:0005634
GO:0046983
GO:0003677
GO:0006355
GO:0006357
GO:0000981
GO:0016021
GO:0035225
GO:0045944
GO:0046982
GO:0007419
GO:0007411
GO:0040011
GO:0007409
GO:0007418
GO:0000978
GO:0043565
GO:0007398
GO:0007420
GO:0007628
GO:0001657
GO:0003824
GO:0008152
GO:0016491
GO:0015930
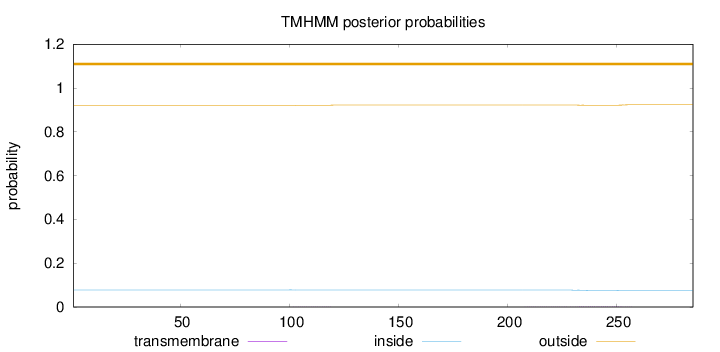
Topology
Subcellular location
Nucleus
Length:
285
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.06044
Exp number, first 60 AAs:
0.00126
Total prob of N-in:
0.07814
outside
1 - 285
Population Genetic Test Statistics
Pi
212.300422
Theta
149.168889
Tajima's D
1.60155
CLR
0.083394
CSRT
0.813359332033398
Interpretation
Uncertain
