Pre Gene Modal
BGIBMGA003822
Annotation
PREDICTED:_protein_mahjong_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.393
Sequence
CDS
ATGGATTCGCCACCGGCTCAAGAGGTAACAGAACTGTTGAGGCAATGGGAGGAACAACATACGCAGCCAAATTACGATCCTATACCCACTTTGACGAGGATAGCAGAGATCATAGAAGCAGAGACAGAGAACTTCATGAAAAAAGACCCAGACCCGTTTGACGAGAGACATCCATCCAGGACAGATCCGGAGTGTGCTCTGGGCCATGCACTCAAAGTTATGTTTAAGAAAGACAATTTTATGACTAAGTTAGTTAATGACTACGTCCGAGACACATACTACTCAAGGCAGAATATCACAGGACGAGATGTCCACAAGTTAAATGTTGCTGCGTGCAGACTGACCTTAGACCTGATGCCCGGATTAGAAATGAGCGTCGTATTTCAGGATAACGAGGCATTGATACAGAGGCTAGTGAACTGGGCGACCAGCTCCACGGAGCCGCTGCAGTGCTACGCCACGGGCCTGCTCGCAGCCGCGATGGAGGTGCAGGATATAGCTACTAATTTTAGGGACCTAAATGCAATGTTAGTTCCTCTCATGTTGAGGAGGCTGCACGGCTTGAGGAACAAAAACCAAGATGATAAGCAATGCAGTCAAGTAACGCCGCCCAGTCAGACGAGACACTTCGCGCGCTTCGACAAGAAACGCAATAACGATATAAAATGCAATGGACCGGGGTCAGAGAAAAATGGAGAACCAGATCGAGATAAAGACAAAGATGGGGGCGCCGAGGACATGGCGGTCGACGATCCGCCCACGACAGATCCCGAGACGCCGGTTAAAAATTACAATAACAGCGCCGCGTCCCCCCCCGCGTGGGGCATGATGTCCCCCCCGCCGCGCCCGCCGCCCCCCTCCGCGCACCACAACCACGAGACCTCCTCCAACTCGAGCTGGGCGGAGATGGAGACCTACGTCATAGGTAACATACAGATACACCCGCCGACCGACGCCACTAAGCAGATGCTCATACTCAGGTACCTGACACCGATGGGAGAATATCAAGAATTTCTGTCACATGTCTTCGAGCAGAACGCCCTAGGGCTGATCCTGGGGTACCTCAACGTACGGGAGTCCCGAGACTCCCGTCTGGCCTTCGAAGCCCTAAAGTACTTGGCGGCGCTCCTGTGCCACAAGAAGTTCTCGATAGACTTTATAAACATGGGAGGGTTGCAGAAATTCTTAGAAGTACCAAGGCCATCCGTCGCCGCCACCGGCGTCTCCATCTGCCTGTACTACCTGGCCTACTGCGAGGACGCAATGGAACGCGTCTGCCTCTTGCCGAGGAAGACTTTAGCGGACTTAGTGCAATACGCCCTGTGGCTTCTAGAATGTTCGCACGACTCCGGCCGCTGCCACGCGACCATGTTCTTCGGCCTCTCGTTCCAGTTCCGGGTCATTCTCGAGGAGTTCGATAACCAGGACGGTCTGAGGAAGCTGTACAACGTGATATCGACGCTCCCGATCCTCGGCTCCGACGAGGAGGAGTCCAGGGTGTCGGACGACGAGACCTGCGCCGCGCGGCAGATCGTCAGGCACGTCTGCGTCGCCCTCAGGAGGTACCTGGAGGCGCACCTCCGGCTAAAGGCGGCGCAGGTGTCCCGGCAGCACGGCGACAGCGTGCCCGACCCGCCACCCTACAAGGTTAGTGGACGTCGTCTCGCCAAGCTTTTAGTTGCCCATGGACATATGTATTGA
Protein
MDSPPAQEVTELLRQWEEQHTQPNYDPIPTLTRIAEIIEAETENFMKKDPDPFDERHPSRTDPECALGHALKVMFKKDNFMTKLVNDYVRDTYYSRQNITGRDVHKLNVAACRLTLDLMPGLEMSVVFQDNEALIQRLVNWATSSTEPLQCYATGLLAAAMEVQDIATNFRDLNAMLVPLMLRRLHGLRNKNQDDKQCSQVTPPSQTRHFARFDKKRNNDIKCNGPGSEKNGEPDRDKDKDGGAEDMAVDDPPTTDPETPVKNYNNSAASPPAWGMMSPPPRPPPPSAHHNHETSSNSSWAEMETYVIGNIQIHPPTDATKQMLILRYLTPMGEYQEFLSHVFEQNALGLILGYLNVRESRDSRLAFEALKYLAALLCHKKFSIDFINMGGLQKFLEVPRPSVAATGVSICLYYLAYCEDAMERVCLLPRKTLADLVQYALWLLECSHDSGRCHATMFFGLSFQFRVILEEFDNQDGLRKLYNVISTLPILGSDEEESRVSDDETCAARQIVRHVCVALRRYLEAHLRLKAAQVSRQHGDSVPDPPPYKVSGRRLAKLLVAHGHMY
Summary
Uniprot
H9J2T5
A0A3S2PEH5
S4PF91
A0A212F5C9
A0A194Q0X0
A0A069DY73
+ More
K7IMJ1 A0A224XHV9 A0A146LRX2 A0A146KJI0 D6WJD4 A0A0P5Z8X9 A0A0P6D1V7 A0A0P5MKG0 A0A0N8AD33 A0A0P5K0R4 A0A0P5JXW4 A0A0N8BGJ6 A0A164KEF0 A0A0P6CVD7 A0A0P5QVM6 A0A0P6E9P6 A0A0P6FZA4 A0A0P5Q4R1 E9GVW2 A0A0P6CHP2 A0A0P5XZB4 A0A0P5N8F4 A0A0P5NAL5 A0A0P5YES2 A0A0P6E2L8 A0A0L7RH82 B0X6E2 W4VRK8 A0A182SUR4 A0A023EWE5 Q17PN7 A0A1S4EVD4 T1J9J0 A0A182KT44 Q7PZX5 A0A182VKI7 A0A182HPP5 A0A131XYM6 A0A182X032 A0A1A9WIG1 A0A182K226 A0A336LYJ1 A0A1S3K840 A0A1S3K7Q1 A0A182WD69 R7VL56 A0A1J1IC47 B4KS40 A0A3Q1BNF8 A0A0Q9XA42 B7PZ89 W5N7K9 A0A3Q3B1R8 B4JWH7 H3B9R8 A0A3B4WWW4 A0A3B4F330 A0A3P8R4X5 A0A0F8AIJ7 A0A3Q3BQQ8 A0A3Q1HXL7 A0A3P9DAF2 C3YPF6 A0A3Q3DWD7 A0A3Q3S4D7 A0A3Q2CXL5 A0A3B4DRT9 I3KLA0 A0A2Z2L0N8 A0A3N0XMC5 A0A087TRG9 A0A3P8YQZ6
K7IMJ1 A0A224XHV9 A0A146LRX2 A0A146KJI0 D6WJD4 A0A0P5Z8X9 A0A0P6D1V7 A0A0P5MKG0 A0A0N8AD33 A0A0P5K0R4 A0A0P5JXW4 A0A0N8BGJ6 A0A164KEF0 A0A0P6CVD7 A0A0P5QVM6 A0A0P6E9P6 A0A0P6FZA4 A0A0P5Q4R1 E9GVW2 A0A0P6CHP2 A0A0P5XZB4 A0A0P5N8F4 A0A0P5NAL5 A0A0P5YES2 A0A0P6E2L8 A0A0L7RH82 B0X6E2 W4VRK8 A0A182SUR4 A0A023EWE5 Q17PN7 A0A1S4EVD4 T1J9J0 A0A182KT44 Q7PZX5 A0A182VKI7 A0A182HPP5 A0A131XYM6 A0A182X032 A0A1A9WIG1 A0A182K226 A0A336LYJ1 A0A1S3K840 A0A1S3K7Q1 A0A182WD69 R7VL56 A0A1J1IC47 B4KS40 A0A3Q1BNF8 A0A0Q9XA42 B7PZ89 W5N7K9 A0A3Q3B1R8 B4JWH7 H3B9R8 A0A3B4WWW4 A0A3B4F330 A0A3P8R4X5 A0A0F8AIJ7 A0A3Q3BQQ8 A0A3Q1HXL7 A0A3P9DAF2 C3YPF6 A0A3Q3DWD7 A0A3Q3S4D7 A0A3Q2CXL5 A0A3B4DRT9 I3KLA0 A0A2Z2L0N8 A0A3N0XMC5 A0A087TRG9 A0A3P8YQZ6
Pubmed
EMBL
BABH01036896
BABH01036897
BABH01036898
RSAL01000072
RVE49076.1
GAIX01004142
+ More
JAA88418.1 AGBW02010216 OWR48909.1 KQ459580 KPI99196.1 GBGD01000098 JAC88791.1 GFTR01008795 JAW07631.1 GDHC01009469 JAQ09160.1 GDHC01021888 JAP96740.1 KQ971343 EFA04458.2 GDIP01047033 JAM56682.1 GDIQ01086381 JAN08356.1 GDIQ01158002 JAK93723.1 GDIP01159273 JAJ64129.1 GDIQ01217095 JAK34630.1 GDIQ01197784 JAK53941.1 GDIQ01179891 JAK71834.1 LRGB01003325 KZS03192.1 GDIQ01089727 JAN05010.1 GDIQ01118417 JAL33309.1 GDIQ01065835 JAN28902.1 GDIQ01051898 JAN42839.1 GDIQ01125662 JAL26064.1 GL732569 EFX76381.1 GDIP01008474 JAM95241.1 GDIP01066329 JAM37386.1 GDIQ01148413 JAL03313.1 GDIQ01148412 JAL03314.1 GDIP01058729 JAM44986.1 GDIQ01068688 JAN26049.1 KQ414592 KOC70220.1 DS232412 EDS41425.1 GANO01002028 JAB57843.1 GAPW01000013 JAC13585.1 CH477190 EAT48714.1 JH431975 AAAB01008986 EAA00204.4 APCN01003267 GEFM01004438 JAP71358.1 UFQT01000229 SSX22081.1 AMQN01016558 KB292190 ELU17976.1 CVRI01000047 CRK97853.1 CH933808 EDW10476.2 KRG05276.1 ABJB010013070 ABJB010083924 ABJB010383163 ABJB010751476 ABJB011073065 DS825131 EEC11911.1 AHAT01015168 CH916375 EDV98315.1 AFYH01041495 AFYH01041496 AFYH01041497 AFYH01041498 AFYH01041499 KQ041821 KKF22034.1 GG666538 EEN57740.1 AERX01002759 KX756656 ASA69199.1 RJVU01069130 ROI74347.1 KK116405 KFM67708.1
JAA88418.1 AGBW02010216 OWR48909.1 KQ459580 KPI99196.1 GBGD01000098 JAC88791.1 GFTR01008795 JAW07631.1 GDHC01009469 JAQ09160.1 GDHC01021888 JAP96740.1 KQ971343 EFA04458.2 GDIP01047033 JAM56682.1 GDIQ01086381 JAN08356.1 GDIQ01158002 JAK93723.1 GDIP01159273 JAJ64129.1 GDIQ01217095 JAK34630.1 GDIQ01197784 JAK53941.1 GDIQ01179891 JAK71834.1 LRGB01003325 KZS03192.1 GDIQ01089727 JAN05010.1 GDIQ01118417 JAL33309.1 GDIQ01065835 JAN28902.1 GDIQ01051898 JAN42839.1 GDIQ01125662 JAL26064.1 GL732569 EFX76381.1 GDIP01008474 JAM95241.1 GDIP01066329 JAM37386.1 GDIQ01148413 JAL03313.1 GDIQ01148412 JAL03314.1 GDIP01058729 JAM44986.1 GDIQ01068688 JAN26049.1 KQ414592 KOC70220.1 DS232412 EDS41425.1 GANO01002028 JAB57843.1 GAPW01000013 JAC13585.1 CH477190 EAT48714.1 JH431975 AAAB01008986 EAA00204.4 APCN01003267 GEFM01004438 JAP71358.1 UFQT01000229 SSX22081.1 AMQN01016558 KB292190 ELU17976.1 CVRI01000047 CRK97853.1 CH933808 EDW10476.2 KRG05276.1 ABJB010013070 ABJB010083924 ABJB010383163 ABJB010751476 ABJB011073065 DS825131 EEC11911.1 AHAT01015168 CH916375 EDV98315.1 AFYH01041495 AFYH01041496 AFYH01041497 AFYH01041498 AFYH01041499 KQ041821 KKF22034.1 GG666538 EEN57740.1 AERX01002759 KX756656 ASA69199.1 RJVU01069130 ROI74347.1 KK116405 KFM67708.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000053268
UP000002358
UP000007266
+ More
UP000076858 UP000000305 UP000053825 UP000002320 UP000075901 UP000008820 UP000075882 UP000007062 UP000075903 UP000075840 UP000076407 UP000091820 UP000075881 UP000085678 UP000075920 UP000014760 UP000183832 UP000009192 UP000257160 UP000001555 UP000018468 UP000264800 UP000001070 UP000008672 UP000261360 UP000261460 UP000265100 UP000264840 UP000265040 UP000265160 UP000001554 UP000264820 UP000261640 UP000265020 UP000261440 UP000005207 UP000054359 UP000265140
UP000076858 UP000000305 UP000053825 UP000002320 UP000075901 UP000008820 UP000075882 UP000007062 UP000075903 UP000075840 UP000076407 UP000091820 UP000075881 UP000085678 UP000075920 UP000014760 UP000183832 UP000009192 UP000257160 UP000001555 UP000018468 UP000264800 UP000001070 UP000008672 UP000261360 UP000261460 UP000265100 UP000264840 UP000265040 UP000265160 UP000001554 UP000264820 UP000261640 UP000265020 UP000261440 UP000005207 UP000054359 UP000265140
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9J2T5
A0A3S2PEH5
S4PF91
A0A212F5C9
A0A194Q0X0
A0A069DY73
+ More
K7IMJ1 A0A224XHV9 A0A146LRX2 A0A146KJI0 D6WJD4 A0A0P5Z8X9 A0A0P6D1V7 A0A0P5MKG0 A0A0N8AD33 A0A0P5K0R4 A0A0P5JXW4 A0A0N8BGJ6 A0A164KEF0 A0A0P6CVD7 A0A0P5QVM6 A0A0P6E9P6 A0A0P6FZA4 A0A0P5Q4R1 E9GVW2 A0A0P6CHP2 A0A0P5XZB4 A0A0P5N8F4 A0A0P5NAL5 A0A0P5YES2 A0A0P6E2L8 A0A0L7RH82 B0X6E2 W4VRK8 A0A182SUR4 A0A023EWE5 Q17PN7 A0A1S4EVD4 T1J9J0 A0A182KT44 Q7PZX5 A0A182VKI7 A0A182HPP5 A0A131XYM6 A0A182X032 A0A1A9WIG1 A0A182K226 A0A336LYJ1 A0A1S3K840 A0A1S3K7Q1 A0A182WD69 R7VL56 A0A1J1IC47 B4KS40 A0A3Q1BNF8 A0A0Q9XA42 B7PZ89 W5N7K9 A0A3Q3B1R8 B4JWH7 H3B9R8 A0A3B4WWW4 A0A3B4F330 A0A3P8R4X5 A0A0F8AIJ7 A0A3Q3BQQ8 A0A3Q1HXL7 A0A3P9DAF2 C3YPF6 A0A3Q3DWD7 A0A3Q3S4D7 A0A3Q2CXL5 A0A3B4DRT9 I3KLA0 A0A2Z2L0N8 A0A3N0XMC5 A0A087TRG9 A0A3P8YQZ6
K7IMJ1 A0A224XHV9 A0A146LRX2 A0A146KJI0 D6WJD4 A0A0P5Z8X9 A0A0P6D1V7 A0A0P5MKG0 A0A0N8AD33 A0A0P5K0R4 A0A0P5JXW4 A0A0N8BGJ6 A0A164KEF0 A0A0P6CVD7 A0A0P5QVM6 A0A0P6E9P6 A0A0P6FZA4 A0A0P5Q4R1 E9GVW2 A0A0P6CHP2 A0A0P5XZB4 A0A0P5N8F4 A0A0P5NAL5 A0A0P5YES2 A0A0P6E2L8 A0A0L7RH82 B0X6E2 W4VRK8 A0A182SUR4 A0A023EWE5 Q17PN7 A0A1S4EVD4 T1J9J0 A0A182KT44 Q7PZX5 A0A182VKI7 A0A182HPP5 A0A131XYM6 A0A182X032 A0A1A9WIG1 A0A182K226 A0A336LYJ1 A0A1S3K840 A0A1S3K7Q1 A0A182WD69 R7VL56 A0A1J1IC47 B4KS40 A0A3Q1BNF8 A0A0Q9XA42 B7PZ89 W5N7K9 A0A3Q3B1R8 B4JWH7 H3B9R8 A0A3B4WWW4 A0A3B4F330 A0A3P8R4X5 A0A0F8AIJ7 A0A3Q3BQQ8 A0A3Q1HXL7 A0A3P9DAF2 C3YPF6 A0A3Q3DWD7 A0A3Q3S4D7 A0A3Q2CXL5 A0A3B4DRT9 I3KLA0 A0A2Z2L0N8 A0A3N0XMC5 A0A087TRG9 A0A3P8YQZ6
Ontologies
GO
PANTHER
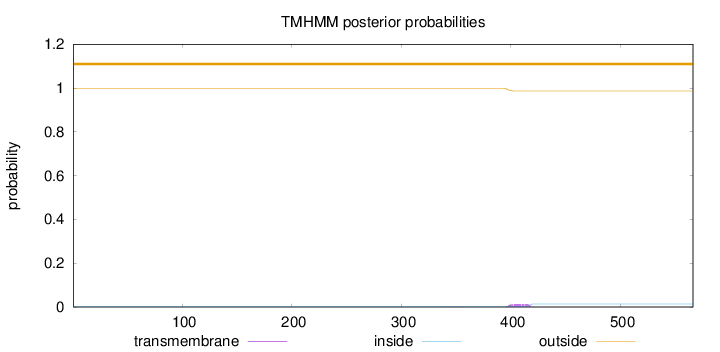
Topology
Length:
566
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.2659
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00079
outside
1 - 566
Population Genetic Test Statistics
Pi
324.821544
Theta
174.323713
Tajima's D
2.201421
CLR
0.132398
CSRT
0.911454427278636
Interpretation
Uncertain
