Gene
KWMTBOMO14742
Pre Gene Modal
BGIBMGA003825
Annotation
PREDICTED:_kinesin-like_protein_KIF3A_isoform_X2_[Bombyx_mori]
Full name
Kinesin-like protein
Location in the cell
Nuclear Reliability : 3.056
Sequence
CDS
ATGCCCGACGCAGAGCTCTCGGCTCCGACCGTCGAGAATGTTCGGGTGGTGGTTCGCGTGCGGCCCATGGACCAGAGGGAGAAAATGGACGGCTCCTACAATTGCGTGTCCGTAGATTCCGTGAACGGGACTGTGGCTGTGACCAGGAACAACGTGACGCCTCCAGAGCCGCCTCGCGTTTATGCTTACGATGCTGTCTTCGACTCTAACACCAGTCAGATGGACATATACGTCCAAACGGCCAGCCCGATTGTCGAGCAAGTCCTCAAAGGGTACAACGGCACTATATTCGCGTACGGGCAGACGGGAACAGGGAAGACCTATACAATGGCGGGCAGCAGCACGGCTCCAGAACTCAGGGGGATCATACCGAACTCGTTTGCACACATCTTCAGTCACATAGCGAAGGCCAAGGACGACGAGAAATTCCTGGTGTGCGTGACTTACCTGGAGATCTACAATGAGGAAGTCCGGGACTTGCTCGGGAACAACCCCCATCAGAGCCTCGAGGTTAAGGAGCGGCCTGACATCGGTGTGTTCGTCAAGGACCTGACAGGGTACGTGGTGCATAACGCTGACGAGCTAGAGAAGATAATGACGGTAGGCGATAAGAACCGGCACATCGGCGCGACCGCCATGAACACGGAGAGCTCGCGGTCCCACGCCATATTCTCGATAACCGTCGAGAGCAGTAAGAAGGGCAGCGACGGTAAAACTCACGTCAAAATGGGGAAACTGCACCTAGTCGATTTGGCGGGGTCGGAGCGTCAAAGCAAGACTCAAGCCACCGGGACACGCCTCAAGGAGGCCACGAAGATAAACCAGTCGCTCTCGGTGCTGGGAAACGTGATATCGGCCCTAGTCGACGGCAAAAGCACCCACATACCGTACAGGAACTCGAAATTGACCAGACTATTGCAGGACTCACTCGGGGGGAATTCTAAGACAGTTATGATCGCGACTATAGGCCCAGCCGATACGAACTACGTCGAGACGATCAGCACGCTTCGTTACGCGAACCGAGCCAAGAATATCGAGAACAAGACGCACGTCAACAACGAACCAGGAGACGCGCTACTTACCAGATTCCAGTTGGAGATCGATCAGCTCAAGAAGCAATTGGAGGACTGTAGCAATGAAATCGAAGGGGGCGCCGAAGATGATTTGTCTGAGGAGGACCTCTCGGAGGACAGCGTGTCCGATCCGGAGATGGACTCTCTCGATCCCGAGGAGAAGAAGCTGCGAAAGAAGATGCGCAGAGAGGAGAAGGAGAAAATGATCAAGGAGAAAACTCATCAGGCCCGGAAAGTTCTGGAGGAGAAGAGAGCTGAACTACAAAGGACTAAGAGACAGCAGGAAGAGTTGAAAGATAAACTGCAGCGTCTCGAGTCTAAGGTTCTGGTAGGCGGAGAGAACCTCCTGGACAAAGCGGACACCCAACGCAGACTCCTGGAGCAGGCCGCCCGGGAGCTCGAGCTCAGGGCCGCCAACGAGCTCAGGCTGAAGGAGGACTTGCAGAAGAAGGAGGCCGAGCGATTGGATATAGAGGAACGCTATTCGAGCCTCCAGGAAGAGAACGCTGCTAAGACCAGGAAGCTCAAGAGGGCTGTCCAACTCCTCAACTCGGCTAAGGCCGAGCTCGCTGACCAGCAGCGCGAGCAGCAGAGGGAGATGGAGGGGATCCTCGACTCGATAAGGGCCCTGAAGAGGGAGATCCAGCTAGCCGATCTCGTGCTGGACTCTTATATACCGAAGGAATATCAGGTTTGTTCGTTGATTTGTCTTTTTATAGCTTAG
Protein
MPDAELSAPTVENVRVVVRVRPMDQREKMDGSYNCVSVDSVNGTVAVTRNNVTPPEPPRVYAYDAVFDSNTSQMDIYVQTASPIVEQVLKGYNGTIFAYGQTGTGKTYTMAGSSTAPELRGIIPNSFAHIFSHIAKAKDDEKFLVCVTYLEIYNEEVRDLLGNNPHQSLEVKERPDIGVFVKDLTGYVVHNADELEKIMTVGDKNRHIGATAMNTESSRSHAIFSITVESSKKGSDGKTHVKMGKLHLVDLAGSERQSKTQATGTRLKEATKINQSLSVLGNVISALVDGKSTHIPYRNSKLTRLLQDSLGGNSKTVMIATIGPADTNYVETISTLRYANRAKNIENKTHVNNEPGDALLTRFQLEIDQLKKQLEDCSNEIEGGAEDDLSEEDLSEDSVSDPEMDSLDPEEKKLRKKMRREEKEKMIKEKTHQARKVLEEKRAELQRTKRQQEELKDKLQRLESKVLVGGENLLDKADTQRRLLEQAARELELRAANELRLKEDLQKKEAERLDIEERYSSLQEENAAKTRKLKRAVQLLNSAKAELADQQREQQREMEGILDSIRALKREIQLADLVLDSYIPKEYQVCSLICLFIA
Summary
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family.
Uniprot
A0A3S2M1V2
A0A194Q1J2
H9J2T8
A0A212F3E5
A0A2W1B6F8
A0A0L0CA29
+ More
Q2LZL5 B4KXE2 A0A3B0JU75 Q9VRK9 A0A0M4E8B0 B3M3R1 A0A0A1WHP1 B4PIS7 B3NGA7 A0A1A9W7E9 A0A1W4UTU5 A0A0J9UEZ6 W8BZ83 A0A154P7T4 A0A0N0U3M8 A0A0C9RKQ3 B4LFR3 A0A2A3EBG7 B4MM15 A0A232EZV1 B7S8Z9 B7S8R3 B4IX13 E0VPH9 A0A067RAN2 A0A1W5A3P4 Q7PQZ7 K7IYI4 A0A087ZX38 S6D9M3 A0A3B4DQL1 A0A3P8WAX8 A0A3Q0RDN9 A0A3Q3VVE5 A0A3Q3ABM6 A0A3Q3F4N1 A0A3Q3GPY0 A0A026WQD7 A0A3P8W7Z6 A0A3Q3LJJ5 A0A195CM39 W5U872 A0A3B5L3K3 A0A151J2C0 A0A3Q1D1Q0 M4A3Y4 A0A1A7X931 A0A3Q1BZU6 A0A3Q3GN87 A0A3P8TEC2 A0A1A8F5R3 B4QRJ6 A0A3P8Z925 A0A1A8BD00 A0A1A8EIJ6 A0A1A7ZIH8 E9QB71 A0A3P8WGA3 Q98TI1 A0A1A8PBF7 A0A1A8LPZ6 A0A3Q4H9J0 A0A2K6TE04 F4WIA9 A0A158NZT6 A0A3B4ERE3 A0A3Q2W9Y9 I3KN69 A0A3P8NYR3 A0A3P9BZU9 A0A1A8JUK5 A0A151WQD7 A0A195BMG4 A0A2R8QGI2 A0A195F3J8 A0A2K6TDW5 A0A091KRN9 A0A1S3GCD0 A0A2K6TDP9 A0A2K5V524 F6SR05 A0A151M6X1 A0A2K6TE36 A0A2T7PNQ0 A0A091QFX3 F7EXQ8 A0A3B3RZ71 D2GVH9 A0A287A7U4 A0A2K6PDD6 A0A093R0U8 A0A091SZ05
Q2LZL5 B4KXE2 A0A3B0JU75 Q9VRK9 A0A0M4E8B0 B3M3R1 A0A0A1WHP1 B4PIS7 B3NGA7 A0A1A9W7E9 A0A1W4UTU5 A0A0J9UEZ6 W8BZ83 A0A154P7T4 A0A0N0U3M8 A0A0C9RKQ3 B4LFR3 A0A2A3EBG7 B4MM15 A0A232EZV1 B7S8Z9 B7S8R3 B4IX13 E0VPH9 A0A067RAN2 A0A1W5A3P4 Q7PQZ7 K7IYI4 A0A087ZX38 S6D9M3 A0A3B4DQL1 A0A3P8WAX8 A0A3Q0RDN9 A0A3Q3VVE5 A0A3Q3ABM6 A0A3Q3F4N1 A0A3Q3GPY0 A0A026WQD7 A0A3P8W7Z6 A0A3Q3LJJ5 A0A195CM39 W5U872 A0A3B5L3K3 A0A151J2C0 A0A3Q1D1Q0 M4A3Y4 A0A1A7X931 A0A3Q1BZU6 A0A3Q3GN87 A0A3P8TEC2 A0A1A8F5R3 B4QRJ6 A0A3P8Z925 A0A1A8BD00 A0A1A8EIJ6 A0A1A7ZIH8 E9QB71 A0A3P8WGA3 Q98TI1 A0A1A8PBF7 A0A1A8LPZ6 A0A3Q4H9J0 A0A2K6TE04 F4WIA9 A0A158NZT6 A0A3B4ERE3 A0A3Q2W9Y9 I3KN69 A0A3P8NYR3 A0A3P9BZU9 A0A1A8JUK5 A0A151WQD7 A0A195BMG4 A0A2R8QGI2 A0A195F3J8 A0A2K6TDW5 A0A091KRN9 A0A1S3GCD0 A0A2K6TDP9 A0A2K5V524 F6SR05 A0A151M6X1 A0A2K6TE36 A0A2T7PNQ0 A0A091QFX3 F7EXQ8 A0A3B3RZ71 D2GVH9 A0A287A7U4 A0A2K6PDD6 A0A093R0U8 A0A091SZ05
Pubmed
26354079
19121390
22118469
28756777
26108605
15632085
+ More
17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25830018 17550304 22936249 24495485 28648823 20566863 24845553 12364791 20075255 23938757 24487278 24508170 30249741 23127152 23542700 25069045 23594743 11432825 25186727 21719571 21347285 19892987 22293439 29240929 20010809 25362486
17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25830018 17550304 22936249 24495485 28648823 20566863 24845553 12364791 20075255 23938757 24487278 24508170 30249741 23127152 23542700 25069045 23594743 11432825 25186727 21719571 21347285 19892987 22293439 29240929 20010809 25362486
EMBL
RSAL01000072
RVE49073.1
KQ459580
KPI99193.1
BABH01036876
BABH01036877
+ More
BABH01036878 BABH01036879 AGBW02010570 OWR48260.1 KZ150638 PZC70425.1 JRES01000692 KNC29107.1 CH379069 EAL29492.1 CH933809 EDW18628.1 OUUW01000002 SPP76916.1 AE014296 AAF50786.1 CP012525 ALC43091.1 CH902618 EDV40354.1 GBXI01015930 JAC98361.1 CM000159 EDW93497.1 CH954178 EDV51008.1 CM002912 KMY97730.1 GAMC01004572 JAC01984.1 KQ434835 KZC07913.1 KQ435883 KOX69778.1 GBYB01013867 JAG83634.1 CH940647 EDW69290.1 KZ288292 PBC29077.1 CH963847 EDW73024.2 NNAY01001420 OXU24036.1 EF710656 ACE75374.1 EF710650 ACE75286.1 CH916366 EDV96319.1 DS235366 EEB15285.1 KK852818 KDR15685.1 AAAB01008859 EAA08081.6 HF586476 CCQ71325.1 KK107139 QOIP01000003 EZA57906.1 RLU24660.1 KQ977580 KYN01773.1 JT407877 AHH38153.1 KQ980404 KYN16180.1 HADW01013196 HADX01008153 SBP14596.1 HAEB01006917 HAEC01016260 SBQ53444.1 CM000363 EDX09307.1 HADZ01000575 SBP64516.1 HAEA01017223 SBQ45704.1 HADY01004147 HAEJ01016730 SBP42632.1 CU179640 AJ311602 CAC33801.1 HAEH01006246 HAEI01002286 SBR78710.1 HAEF01008430 HAEG01004376 SBR46588.1 GL888175 EGI66040.1 ADTU01004997 AERX01014108 AERX01014109 AERX01014110 HAED01016289 HAEE01003049 SBR23069.1 KQ982829 KYQ50109.1 KQ976440 KYM86476.1 KQ981856 KYN34659.1 KK753747 KFP42647.1 AQIA01060497 AQIA01060498 AQIA01060499 AKHW03006437 KYO20249.1 PZQS01000003 PVD34987.1 KK696638 KFQ24752.1 GL192350 EFB24258.1 AEMK02000012 KL424111 KFW89792.1 KK492628 KFQ63957.1
BABH01036878 BABH01036879 AGBW02010570 OWR48260.1 KZ150638 PZC70425.1 JRES01000692 KNC29107.1 CH379069 EAL29492.1 CH933809 EDW18628.1 OUUW01000002 SPP76916.1 AE014296 AAF50786.1 CP012525 ALC43091.1 CH902618 EDV40354.1 GBXI01015930 JAC98361.1 CM000159 EDW93497.1 CH954178 EDV51008.1 CM002912 KMY97730.1 GAMC01004572 JAC01984.1 KQ434835 KZC07913.1 KQ435883 KOX69778.1 GBYB01013867 JAG83634.1 CH940647 EDW69290.1 KZ288292 PBC29077.1 CH963847 EDW73024.2 NNAY01001420 OXU24036.1 EF710656 ACE75374.1 EF710650 ACE75286.1 CH916366 EDV96319.1 DS235366 EEB15285.1 KK852818 KDR15685.1 AAAB01008859 EAA08081.6 HF586476 CCQ71325.1 KK107139 QOIP01000003 EZA57906.1 RLU24660.1 KQ977580 KYN01773.1 JT407877 AHH38153.1 KQ980404 KYN16180.1 HADW01013196 HADX01008153 SBP14596.1 HAEB01006917 HAEC01016260 SBQ53444.1 CM000363 EDX09307.1 HADZ01000575 SBP64516.1 HAEA01017223 SBQ45704.1 HADY01004147 HAEJ01016730 SBP42632.1 CU179640 AJ311602 CAC33801.1 HAEH01006246 HAEI01002286 SBR78710.1 HAEF01008430 HAEG01004376 SBR46588.1 GL888175 EGI66040.1 ADTU01004997 AERX01014108 AERX01014109 AERX01014110 HAED01016289 HAEE01003049 SBR23069.1 KQ982829 KYQ50109.1 KQ976440 KYM86476.1 KQ981856 KYN34659.1 KK753747 KFP42647.1 AQIA01060497 AQIA01060498 AQIA01060499 AKHW03006437 KYO20249.1 PZQS01000003 PVD34987.1 KK696638 KFQ24752.1 GL192350 EFB24258.1 AEMK02000012 KL424111 KFW89792.1 KK492628 KFQ63957.1
Proteomes
UP000283053
UP000053268
UP000005204
UP000007151
UP000037069
UP000001819
+ More
UP000009192 UP000268350 UP000000803 UP000092553 UP000007801 UP000002282 UP000008711 UP000091820 UP000192221 UP000076502 UP000053105 UP000008792 UP000242457 UP000007798 UP000215335 UP000001070 UP000009046 UP000027135 UP000192224 UP000007062 UP000002358 UP000005203 UP000261440 UP000265120 UP000261340 UP000261620 UP000264800 UP000261660 UP000053097 UP000279307 UP000261640 UP000078542 UP000221080 UP000261380 UP000078492 UP000257160 UP000002852 UP000265080 UP000000304 UP000265140 UP000000437 UP000261580 UP000233220 UP000007755 UP000005205 UP000261460 UP000264840 UP000005207 UP000265100 UP000265160 UP000075809 UP000078540 UP000078541 UP000081671 UP000233100 UP000002281 UP000050525 UP000245119 UP000008225 UP000261540 UP000008227 UP000233200
UP000009192 UP000268350 UP000000803 UP000092553 UP000007801 UP000002282 UP000008711 UP000091820 UP000192221 UP000076502 UP000053105 UP000008792 UP000242457 UP000007798 UP000215335 UP000001070 UP000009046 UP000027135 UP000192224 UP000007062 UP000002358 UP000005203 UP000261440 UP000265120 UP000261340 UP000261620 UP000264800 UP000261660 UP000053097 UP000279307 UP000261640 UP000078542 UP000221080 UP000261380 UP000078492 UP000257160 UP000002852 UP000265080 UP000000304 UP000265140 UP000000437 UP000261580 UP000233220 UP000007755 UP000005205 UP000261460 UP000264840 UP000005207 UP000265100 UP000265160 UP000075809 UP000078540 UP000078541 UP000081671 UP000233100 UP000002281 UP000050525 UP000245119 UP000008225 UP000261540 UP000008227 UP000233200
Interpro
SUPFAM
SSF52540
SSF52540
Gene 3D
ProteinModelPortal
A0A3S2M1V2
A0A194Q1J2
H9J2T8
A0A212F3E5
A0A2W1B6F8
A0A0L0CA29
+ More
Q2LZL5 B4KXE2 A0A3B0JU75 Q9VRK9 A0A0M4E8B0 B3M3R1 A0A0A1WHP1 B4PIS7 B3NGA7 A0A1A9W7E9 A0A1W4UTU5 A0A0J9UEZ6 W8BZ83 A0A154P7T4 A0A0N0U3M8 A0A0C9RKQ3 B4LFR3 A0A2A3EBG7 B4MM15 A0A232EZV1 B7S8Z9 B7S8R3 B4IX13 E0VPH9 A0A067RAN2 A0A1W5A3P4 Q7PQZ7 K7IYI4 A0A087ZX38 S6D9M3 A0A3B4DQL1 A0A3P8WAX8 A0A3Q0RDN9 A0A3Q3VVE5 A0A3Q3ABM6 A0A3Q3F4N1 A0A3Q3GPY0 A0A026WQD7 A0A3P8W7Z6 A0A3Q3LJJ5 A0A195CM39 W5U872 A0A3B5L3K3 A0A151J2C0 A0A3Q1D1Q0 M4A3Y4 A0A1A7X931 A0A3Q1BZU6 A0A3Q3GN87 A0A3P8TEC2 A0A1A8F5R3 B4QRJ6 A0A3P8Z925 A0A1A8BD00 A0A1A8EIJ6 A0A1A7ZIH8 E9QB71 A0A3P8WGA3 Q98TI1 A0A1A8PBF7 A0A1A8LPZ6 A0A3Q4H9J0 A0A2K6TE04 F4WIA9 A0A158NZT6 A0A3B4ERE3 A0A3Q2W9Y9 I3KN69 A0A3P8NYR3 A0A3P9BZU9 A0A1A8JUK5 A0A151WQD7 A0A195BMG4 A0A2R8QGI2 A0A195F3J8 A0A2K6TDW5 A0A091KRN9 A0A1S3GCD0 A0A2K6TDP9 A0A2K5V524 F6SR05 A0A151M6X1 A0A2K6TE36 A0A2T7PNQ0 A0A091QFX3 F7EXQ8 A0A3B3RZ71 D2GVH9 A0A287A7U4 A0A2K6PDD6 A0A093R0U8 A0A091SZ05
Q2LZL5 B4KXE2 A0A3B0JU75 Q9VRK9 A0A0M4E8B0 B3M3R1 A0A0A1WHP1 B4PIS7 B3NGA7 A0A1A9W7E9 A0A1W4UTU5 A0A0J9UEZ6 W8BZ83 A0A154P7T4 A0A0N0U3M8 A0A0C9RKQ3 B4LFR3 A0A2A3EBG7 B4MM15 A0A232EZV1 B7S8Z9 B7S8R3 B4IX13 E0VPH9 A0A067RAN2 A0A1W5A3P4 Q7PQZ7 K7IYI4 A0A087ZX38 S6D9M3 A0A3B4DQL1 A0A3P8WAX8 A0A3Q0RDN9 A0A3Q3VVE5 A0A3Q3ABM6 A0A3Q3F4N1 A0A3Q3GPY0 A0A026WQD7 A0A3P8W7Z6 A0A3Q3LJJ5 A0A195CM39 W5U872 A0A3B5L3K3 A0A151J2C0 A0A3Q1D1Q0 M4A3Y4 A0A1A7X931 A0A3Q1BZU6 A0A3Q3GN87 A0A3P8TEC2 A0A1A8F5R3 B4QRJ6 A0A3P8Z925 A0A1A8BD00 A0A1A8EIJ6 A0A1A7ZIH8 E9QB71 A0A3P8WGA3 Q98TI1 A0A1A8PBF7 A0A1A8LPZ6 A0A3Q4H9J0 A0A2K6TE04 F4WIA9 A0A158NZT6 A0A3B4ERE3 A0A3Q2W9Y9 I3KN69 A0A3P8NYR3 A0A3P9BZU9 A0A1A8JUK5 A0A151WQD7 A0A195BMG4 A0A2R8QGI2 A0A195F3J8 A0A2K6TDW5 A0A091KRN9 A0A1S3GCD0 A0A2K6TDP9 A0A2K5V524 F6SR05 A0A151M6X1 A0A2K6TE36 A0A2T7PNQ0 A0A091QFX3 F7EXQ8 A0A3B3RZ71 D2GVH9 A0A287A7U4 A0A2K6PDD6 A0A093R0U8 A0A091SZ05
PDB
3B6U
E-value=6.66555e-112,
Score=1035
Ontologies
GO
GO:0005874
GO:0005524
GO:0003774
GO:0007018
GO:0008017
GO:0003777
GO:0005929
GO:0007411
GO:1904115
GO:0005769
GO:0030951
GO:0005886
GO:1905515
GO:0090263
GO:0008089
GO:0007608
GO:0007605
GO:0005912
GO:0005871
GO:0030054
GO:0035845
GO:0046907
GO:0060271
GO:0016887
GO:0009952
GO:0001822
GO:0005814
GO:0010457
GO:0120170
GO:0008544
GO:0034454
GO:0050679
GO:0016939
GO:0021904
GO:0001701
GO:0060122
GO:0036334
GO:0001947
GO:0032391
GO:0061351
GO:0044458
GO:0021542
GO:0031514
GO:0007224
GO:0006886
GO:0005515
GO:0003824
GO:0008152
GO:0016491
GO:0015930
PANTHER
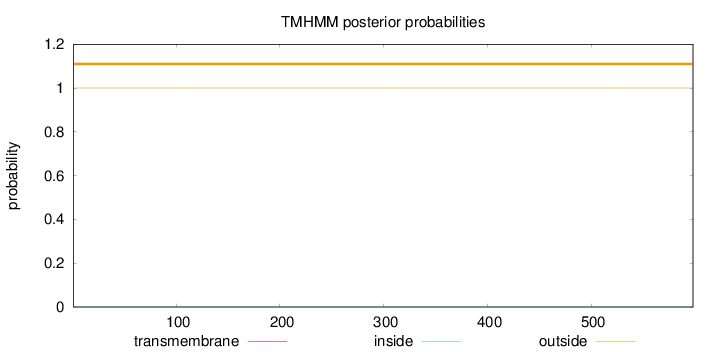
Topology
Length:
598
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00144
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.00023
outside
1 - 598
Population Genetic Test Statistics
Pi
252.035524
Theta
183.042584
Tajima's D
1.420894
CLR
0.109377
CSRT
0.770861456927154
Interpretation
Uncertain
