Gene
KWMTBOMO14741
Pre Gene Modal
BGIBMGA003825
Annotation
PREDICTED:_kinesin-like_protein_KIF3A_isoform_X2_[Bombyx_mori]
Full name
Kinesin-like protein
Location in the cell
Nuclear Reliability : 1.898
Sequence
CDS
ATGGTATTGCATGGAAGTCAAACTAAGACTAATCGAATCGTGTCCGAATTACGGATATATATATATATATCAGTCTGCAATCTGATTATCGATGCTCCTCAGGCCCTGATCGACAAGTACGTGCACTGGAACGAGCAGCTTGGCGAGTGGCAGGTTCGGTGCGTGGCCTACACCGGGAACAACATGAGTTGTCCATTGCAGCCGCGACACGCCGCCACCAACAAACACATGGAGCAACCAGACCTATCAGACAGGTACCTGTCGTACGCAACGGTGAACAGCCGCGGCACCTCACGGCTCGCGCGACCTCATTCATCCGCCGTGCCCCGACCTCACACCGCTTTGAGACAGCGACAGTAG
Protein
MVLHGSQTKTNRIVSELRIYIYISVCNLIIDAPQALIDKYVHWNEQLGEWQVRCVAYTGNNMSCPLQPRHAATNKHMEQPDLSDRYLSYATVNSRGTSRLARPHSSAVPRPHTALRQRQ
Summary
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family.
Feature
chain Kinesin-like protein
Uniprot
A0A2A4IX31
A0A2H1WHC3
A0A3S2M1V2
A0A212F3E9
A0A212F3E5
A0A194R504
+ More
A0A194Q1J2 B4KXE2 B4H3B4 A0A0K8VZE1 T1PKF4 B4LFR3 Q2LZL5 A0A0A1WHP1 A0A3B0JU75 B4HUE8 Q9VRK9 B4PIS7 B3NGA7 A0A1W4UTU5 A0A0M4E8B0 W8BZ83 A0A336MAZ4 A0A336MB45 A0A0J9UEZ6 B4QRJ6 B3M3R1 A0A034VXP7 A0A2J7R1Z9 A0A0L0CA29 B4IX13 B4MM15 A0A1B0CKE8 A0A1I8P160 A0A154P7T4 A0A2M4CS12 A0A182FPI4 W5J6N6 A0A182LF86 A0A0N0U3M8 A0A182HT43 A0A182VBX8 A0A1A9VJ47 A0A182P728 Q7PQZ7 A0A1B0FCA3 A0A182XI59 A0A1A9XV50 A0A1B0BHC8 A0A1B0AEW8 E2B6U2 A0A182JKT3 A0A1S3RQP6 A0A182R9W8 A0A0S7F3B1 A0A1S3RP91 A0A2A3EBG7 A0A182QMI4 A0A087ZX38 A0A182XX09 A0A182SQK3 A0A182VX94 A0A182LTF6 A0A1S3QGX7 A0A1A9W7E9 A0A1V4KTZ3 A0A182NFG0 E1ZXK1 A0A182G831 B0W3J5 A0A1Q3G2R5 A0A1Q3G2W0 Q16RI3 Q6UIP4 Q6UIP3 A0A158NZT6 F4WIA9 A0A151WQD7 A0A026WQD7 A0A151J2C0 A0A195BMG4 A0A1S3RP90 A0A0L7R925 A0A1S3RQZ5 A0A3B5ASV5 K7IYI4 A0A0C9RKQ3 S6D9M3 A0A232EZV1 A0A195F3J8 G3Q2I5 A0A3Q3ABM6 A0A195CM39 A0A3Q1F2U7 A0A067RAN2 A0A3Q3F4N1 A0A3Q3LJJ5 A0A3P8W7Z6
A0A194Q1J2 B4KXE2 B4H3B4 A0A0K8VZE1 T1PKF4 B4LFR3 Q2LZL5 A0A0A1WHP1 A0A3B0JU75 B4HUE8 Q9VRK9 B4PIS7 B3NGA7 A0A1W4UTU5 A0A0M4E8B0 W8BZ83 A0A336MAZ4 A0A336MB45 A0A0J9UEZ6 B4QRJ6 B3M3R1 A0A034VXP7 A0A2J7R1Z9 A0A0L0CA29 B4IX13 B4MM15 A0A1B0CKE8 A0A1I8P160 A0A154P7T4 A0A2M4CS12 A0A182FPI4 W5J6N6 A0A182LF86 A0A0N0U3M8 A0A182HT43 A0A182VBX8 A0A1A9VJ47 A0A182P728 Q7PQZ7 A0A1B0FCA3 A0A182XI59 A0A1A9XV50 A0A1B0BHC8 A0A1B0AEW8 E2B6U2 A0A182JKT3 A0A1S3RQP6 A0A182R9W8 A0A0S7F3B1 A0A1S3RP91 A0A2A3EBG7 A0A182QMI4 A0A087ZX38 A0A182XX09 A0A182SQK3 A0A182VX94 A0A182LTF6 A0A1S3QGX7 A0A1A9W7E9 A0A1V4KTZ3 A0A182NFG0 E1ZXK1 A0A182G831 B0W3J5 A0A1Q3G2R5 A0A1Q3G2W0 Q16RI3 Q6UIP4 Q6UIP3 A0A158NZT6 F4WIA9 A0A151WQD7 A0A026WQD7 A0A151J2C0 A0A195BMG4 A0A1S3RP90 A0A0L7R925 A0A1S3RQZ5 A0A3B5ASV5 K7IYI4 A0A0C9RKQ3 S6D9M3 A0A232EZV1 A0A195F3J8 G3Q2I5 A0A3Q3ABM6 A0A195CM39 A0A3Q1F2U7 A0A067RAN2 A0A3Q3F4N1 A0A3Q3LJJ5 A0A3P8W7Z6
Pubmed
22118469
26354079
17994087
25315136
15632085
25830018
+ More
10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 24495485 22936249 25348373 26108605 20920257 23761445 20966253 12364791 20798317 25244985 26483478 17510324 14557539 21347285 21719571 24508170 30249741 20075255 23938757 28648823 24845553 24487278
10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 24495485 22936249 25348373 26108605 20920257 23761445 20966253 12364791 20798317 25244985 26483478 17510324 14557539 21347285 21719571 24508170 30249741 20075255 23938757 28648823 24845553 24487278
EMBL
NWSH01005009
PCG64497.1
ODYU01008685
SOQ52471.1
RSAL01000072
RVE49073.1
+ More
AGBW02010570 OWR48261.1 OWR48260.1 KQ460685 KPJ12762.1 KQ459580 KPI99193.1 CH933809 EDW18628.1 CH479206 EDW30854.1 GDHF01008087 JAI44227.1 KA648560 AFP63189.1 CH940647 EDW69290.1 CH379069 EAL29492.1 GBXI01015930 JAC98361.1 OUUW01000002 SPP76916.1 CH480817 EDW50569.1 AE014296 AAF50786.1 CM000159 EDW93497.1 CH954178 EDV51008.1 CP012525 ALC43091.1 GAMC01004572 JAC01984.1 UFQT01000835 SSX27504.1 SSX27505.1 CM002912 KMY97730.1 CM000363 EDX09307.1 CH902618 EDV40354.1 GAKP01012312 JAC46640.1 NEVH01008202 PNF34839.1 JRES01000692 KNC29107.1 CH916366 EDV96319.1 CH963847 EDW73024.2 AJWK01016155 KQ434835 KZC07913.1 GGFL01003954 MBW68132.1 ADMH02002069 ETN59656.1 KQ435883 KOX69778.1 APCN01000411 AAAB01008859 EAA08081.6 CCAG010008817 JXJN01014265 GL446009 EFN88583.1 GBYX01471734 JAO09920.1 KZ288292 PBC29077.1 AXCN02000214 AXCM01008924 LSYS01001584 OPJ87914.1 GL435066 EFN74084.1 JXUM01047181 KQ561509 KXJ78351.1 DS231832 EDS31781.1 GFDL01000953 JAV34092.1 GFDL01000966 JAV34079.1 CH477709 EAT37007.1 AY369822 AAR11251.1 AY369823 AAR11252.1 ADTU01004997 GL888175 EGI66040.1 KQ982829 KYQ50109.1 KK107139 QOIP01000003 EZA57906.1 RLU24660.1 KQ980404 KYN16180.1 KQ976440 KYM86476.1 KQ414628 KOC67367.1 GBYB01013867 JAG83634.1 HF586476 CCQ71325.1 NNAY01001420 OXU24036.1 KQ981856 KYN34659.1 KQ977580 KYN01773.1 KK852818 KDR15685.1
AGBW02010570 OWR48261.1 OWR48260.1 KQ460685 KPJ12762.1 KQ459580 KPI99193.1 CH933809 EDW18628.1 CH479206 EDW30854.1 GDHF01008087 JAI44227.1 KA648560 AFP63189.1 CH940647 EDW69290.1 CH379069 EAL29492.1 GBXI01015930 JAC98361.1 OUUW01000002 SPP76916.1 CH480817 EDW50569.1 AE014296 AAF50786.1 CM000159 EDW93497.1 CH954178 EDV51008.1 CP012525 ALC43091.1 GAMC01004572 JAC01984.1 UFQT01000835 SSX27504.1 SSX27505.1 CM002912 KMY97730.1 CM000363 EDX09307.1 CH902618 EDV40354.1 GAKP01012312 JAC46640.1 NEVH01008202 PNF34839.1 JRES01000692 KNC29107.1 CH916366 EDV96319.1 CH963847 EDW73024.2 AJWK01016155 KQ434835 KZC07913.1 GGFL01003954 MBW68132.1 ADMH02002069 ETN59656.1 KQ435883 KOX69778.1 APCN01000411 AAAB01008859 EAA08081.6 CCAG010008817 JXJN01014265 GL446009 EFN88583.1 GBYX01471734 JAO09920.1 KZ288292 PBC29077.1 AXCN02000214 AXCM01008924 LSYS01001584 OPJ87914.1 GL435066 EFN74084.1 JXUM01047181 KQ561509 KXJ78351.1 DS231832 EDS31781.1 GFDL01000953 JAV34092.1 GFDL01000966 JAV34079.1 CH477709 EAT37007.1 AY369822 AAR11251.1 AY369823 AAR11252.1 ADTU01004997 GL888175 EGI66040.1 KQ982829 KYQ50109.1 KK107139 QOIP01000003 EZA57906.1 RLU24660.1 KQ980404 KYN16180.1 KQ976440 KYM86476.1 KQ414628 KOC67367.1 GBYB01013867 JAG83634.1 HF586476 CCQ71325.1 NNAY01001420 OXU24036.1 KQ981856 KYN34659.1 KQ977580 KYN01773.1 KK852818 KDR15685.1
Proteomes
UP000218220
UP000283053
UP000007151
UP000053240
UP000053268
UP000009192
+ More
UP000008744 UP000095301 UP000008792 UP000001819 UP000268350 UP000001292 UP000000803 UP000002282 UP000008711 UP000192221 UP000092553 UP000000304 UP000007801 UP000235965 UP000037069 UP000001070 UP000007798 UP000092461 UP000095300 UP000076502 UP000069272 UP000000673 UP000075882 UP000053105 UP000075840 UP000075903 UP000078200 UP000075885 UP000007062 UP000092444 UP000076407 UP000092443 UP000092460 UP000092445 UP000008237 UP000075880 UP000087266 UP000075900 UP000242457 UP000075886 UP000005203 UP000076408 UP000075901 UP000075920 UP000075883 UP000091820 UP000190648 UP000075884 UP000000311 UP000069940 UP000249989 UP000002320 UP000008820 UP000005205 UP000007755 UP000075809 UP000053097 UP000279307 UP000078492 UP000078540 UP000053825 UP000261400 UP000002358 UP000215335 UP000078541 UP000007635 UP000264800 UP000078542 UP000257200 UP000027135 UP000261640 UP000265120
UP000008744 UP000095301 UP000008792 UP000001819 UP000268350 UP000001292 UP000000803 UP000002282 UP000008711 UP000192221 UP000092553 UP000000304 UP000007801 UP000235965 UP000037069 UP000001070 UP000007798 UP000092461 UP000095300 UP000076502 UP000069272 UP000000673 UP000075882 UP000053105 UP000075840 UP000075903 UP000078200 UP000075885 UP000007062 UP000092444 UP000076407 UP000092443 UP000092460 UP000092445 UP000008237 UP000075880 UP000087266 UP000075900 UP000242457 UP000075886 UP000005203 UP000076408 UP000075901 UP000075920 UP000075883 UP000091820 UP000190648 UP000075884 UP000000311 UP000069940 UP000249989 UP000002320 UP000008820 UP000005205 UP000007755 UP000075809 UP000053097 UP000279307 UP000078492 UP000078540 UP000053825 UP000261400 UP000002358 UP000215335 UP000078541 UP000007635 UP000264800 UP000078542 UP000257200 UP000027135 UP000261640 UP000265120
PRIDE
Interpro
SUPFAM
SSF52540
SSF52540
Gene 3D
ProteinModelPortal
A0A2A4IX31
A0A2H1WHC3
A0A3S2M1V2
A0A212F3E9
A0A212F3E5
A0A194R504
+ More
A0A194Q1J2 B4KXE2 B4H3B4 A0A0K8VZE1 T1PKF4 B4LFR3 Q2LZL5 A0A0A1WHP1 A0A3B0JU75 B4HUE8 Q9VRK9 B4PIS7 B3NGA7 A0A1W4UTU5 A0A0M4E8B0 W8BZ83 A0A336MAZ4 A0A336MB45 A0A0J9UEZ6 B4QRJ6 B3M3R1 A0A034VXP7 A0A2J7R1Z9 A0A0L0CA29 B4IX13 B4MM15 A0A1B0CKE8 A0A1I8P160 A0A154P7T4 A0A2M4CS12 A0A182FPI4 W5J6N6 A0A182LF86 A0A0N0U3M8 A0A182HT43 A0A182VBX8 A0A1A9VJ47 A0A182P728 Q7PQZ7 A0A1B0FCA3 A0A182XI59 A0A1A9XV50 A0A1B0BHC8 A0A1B0AEW8 E2B6U2 A0A182JKT3 A0A1S3RQP6 A0A182R9W8 A0A0S7F3B1 A0A1S3RP91 A0A2A3EBG7 A0A182QMI4 A0A087ZX38 A0A182XX09 A0A182SQK3 A0A182VX94 A0A182LTF6 A0A1S3QGX7 A0A1A9W7E9 A0A1V4KTZ3 A0A182NFG0 E1ZXK1 A0A182G831 B0W3J5 A0A1Q3G2R5 A0A1Q3G2W0 Q16RI3 Q6UIP4 Q6UIP3 A0A158NZT6 F4WIA9 A0A151WQD7 A0A026WQD7 A0A151J2C0 A0A195BMG4 A0A1S3RP90 A0A0L7R925 A0A1S3RQZ5 A0A3B5ASV5 K7IYI4 A0A0C9RKQ3 S6D9M3 A0A232EZV1 A0A195F3J8 G3Q2I5 A0A3Q3ABM6 A0A195CM39 A0A3Q1F2U7 A0A067RAN2 A0A3Q3F4N1 A0A3Q3LJJ5 A0A3P8W7Z6
A0A194Q1J2 B4KXE2 B4H3B4 A0A0K8VZE1 T1PKF4 B4LFR3 Q2LZL5 A0A0A1WHP1 A0A3B0JU75 B4HUE8 Q9VRK9 B4PIS7 B3NGA7 A0A1W4UTU5 A0A0M4E8B0 W8BZ83 A0A336MAZ4 A0A336MB45 A0A0J9UEZ6 B4QRJ6 B3M3R1 A0A034VXP7 A0A2J7R1Z9 A0A0L0CA29 B4IX13 B4MM15 A0A1B0CKE8 A0A1I8P160 A0A154P7T4 A0A2M4CS12 A0A182FPI4 W5J6N6 A0A182LF86 A0A0N0U3M8 A0A182HT43 A0A182VBX8 A0A1A9VJ47 A0A182P728 Q7PQZ7 A0A1B0FCA3 A0A182XI59 A0A1A9XV50 A0A1B0BHC8 A0A1B0AEW8 E2B6U2 A0A182JKT3 A0A1S3RQP6 A0A182R9W8 A0A0S7F3B1 A0A1S3RP91 A0A2A3EBG7 A0A182QMI4 A0A087ZX38 A0A182XX09 A0A182SQK3 A0A182VX94 A0A182LTF6 A0A1S3QGX7 A0A1A9W7E9 A0A1V4KTZ3 A0A182NFG0 E1ZXK1 A0A182G831 B0W3J5 A0A1Q3G2R5 A0A1Q3G2W0 Q16RI3 Q6UIP4 Q6UIP3 A0A158NZT6 F4WIA9 A0A151WQD7 A0A026WQD7 A0A151J2C0 A0A195BMG4 A0A1S3RP90 A0A0L7R925 A0A1S3RQZ5 A0A3B5ASV5 K7IYI4 A0A0C9RKQ3 S6D9M3 A0A232EZV1 A0A195F3J8 G3Q2I5 A0A3Q3ABM6 A0A195CM39 A0A3Q1F2U7 A0A067RAN2 A0A3Q3F4N1 A0A3Q3LJJ5 A0A3P8W7Z6
Ontologies
GO
PANTHER
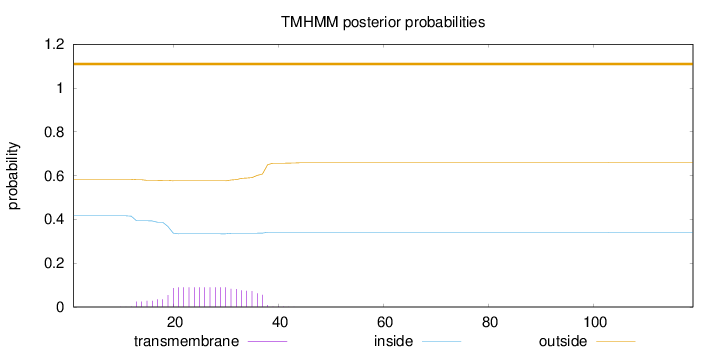
Topology
Length:
119
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.73924
Exp number, first 60 AAs:
1.73907
Total prob of N-in:
0.41623
outside
1 - 119
Population Genetic Test Statistics
Pi
262.127827
Theta
171.825441
Tajima's D
1.740222
CLR
0.160735
CSRT
0.838458077096145
Interpretation
Uncertain
