Gene
KWMTBOMO14737
Pre Gene Modal
BGIBMGA003827
Annotation
PREDICTED:_ubiquitin-associated_domain-containing_protein_1_isoform_X1_[Bombyx_mori]
Full name
Peptidyl-prolyl cis-trans isomerase
+ More
Peptide hydrolase
Peptide hydrolase
Location in the cell
Extracellular Reliability : 1.491
Sequence
CDS
ATGTCAATTTATGTCCCAGGTCTGTACTGGAGTGGAGGAGACGTGATACACGACAATGGCTTCGGCTGTTACGCCCAAAGGGGAAGACGGACGCCCATCGGAGCCGAGAACTATCACTACTCACATTCTCTTCCCGGTCTTCTGTCGATGCGCGTGACAAAGGACGATGAAATGTGCGGCATATTTAATATAACCTTCAAACCGCTGCCTCAATTCGATCTACGGAACGTTGTCTTTGGAAGAGTAATTCTGCCGTGCGAGACTTTTGAAGTGATTCGCGGCTTAGGCGCCCCGCTCAGCTCGCGTCCGGTCGTGGAGATATCGGCGGCAAAACACCGCGTCGACAACCGCTGGGTGTACGGAAAGCCTAATAGTAGAATTAAAAACTGA
Protein
MSIYVPGLYWSGGDVIHDNGFGCYAQRGRRTPIGAENYHYSHSLPGLLSMRVTKDDEMCGIFNITFKPLPQFDLRNVVFGRVILPCETFEVIRGLGAPLSSRPVVEISAAKHRVDNRWVYGKPNSRIKN
Summary
Description
PPIases accelerate the folding of proteins. It catalyzes the cis-trans isomerization of proline imidic peptide bonds in oligopeptides.
Catalytic Activity
[protein]-peptidylproline (omega=180) = [protein]-peptidylproline (omega=0)
Similarity
Belongs to the cyclophilin-type PPIase family.
Belongs to the peptidase M28 family.
Belongs to the peptidase M28 family.
Feature
chain Peptidyl-prolyl cis-trans isomerase
Uniprot
A0A2H1V7R5
A0A2A4IYK9
A0A212F543
A0A194Q0T1
H9IV42
Q2F611
+ More
A0A212EX06 A0A2A4JK20 A0A194RBU3 A0A3S2NYK6 A0A194QC44 A0A2H1VVT0 J9KSD8 A0A0J0XHS7 A0A099P773 A0A1Q2YAJ4 D8MAZ2 A0A1R2BFH5 L7CGQ4 Q7UMR2 K5D633 F2AX20 F2UN80 M5SCB6 M5TDW4 C5M6A0 M2AZF2 A0A1E7F4I6 A0A132A4N0 W0E0J2 A0A1B7SM77 M0SV84 A0A3M8NRN9 A0A1Y1HSP4 H2AUQ6 L1IT53 A0A090LEW7 A0A162B4Q9 F2QQD5 A0A1Q3A3R1 C5DYK2 W7TPH9 H3A5C5 A5K7K7 A0A0J9TWG5 A0A0J9W0E8 A0A0J9SG55 A0A1G4HBM6 A0A1B2JFQ6 A0A376B2U8 A0A0J9SVE3 A0A0J1B9C4 D8TPQ4 A0A1E3NGA1 A0A1E4TA10 M0U4B7 M5UGQ2 C3YR41 A0A061AWC1 K0KUU1 A0A1E3NW22 A0A287I203 A5KDH9 A0A1B1E4J5 I2JRN0 A0A0V1Q606 A0A1V2L5F3 A0A0B7FXH5 A5K4H1 A0A0J9TXW6 A0A0J9TCI7 A0A0J9V388 A0A1G4HCM3 H8Z5U6 A0A0D9WU02 A0A0N5BJH8 A0A1R2C8K2 A0A2N1J8S6 A0A1X7R6L9 A0A1J3JAS4 V7CSY1 M1UUR5 A0A165FND5 A0A165FP01
A0A212EX06 A0A2A4JK20 A0A194RBU3 A0A3S2NYK6 A0A194QC44 A0A2H1VVT0 J9KSD8 A0A0J0XHS7 A0A099P773 A0A1Q2YAJ4 D8MAZ2 A0A1R2BFH5 L7CGQ4 Q7UMR2 K5D633 F2AX20 F2UN80 M5SCB6 M5TDW4 C5M6A0 M2AZF2 A0A1E7F4I6 A0A132A4N0 W0E0J2 A0A1B7SM77 M0SV84 A0A3M8NRN9 A0A1Y1HSP4 H2AUQ6 L1IT53 A0A090LEW7 A0A162B4Q9 F2QQD5 A0A1Q3A3R1 C5DYK2 W7TPH9 H3A5C5 A5K7K7 A0A0J9TWG5 A0A0J9W0E8 A0A0J9SG55 A0A1G4HBM6 A0A1B2JFQ6 A0A376B2U8 A0A0J9SVE3 A0A0J1B9C4 D8TPQ4 A0A1E3NGA1 A0A1E4TA10 M0U4B7 M5UGQ2 C3YR41 A0A061AWC1 K0KUU1 A0A1E3NW22 A0A287I203 A5KDH9 A0A1B1E4J5 I2JRN0 A0A0V1Q606 A0A1V2L5F3 A0A0B7FXH5 A5K4H1 A0A0J9TXW6 A0A0J9TCI7 A0A0J9V388 A0A1G4HCM3 H8Z5U6 A0A0D9WU02 A0A0N5BJH8 A0A1R2C8K2 A0A2N1J8S6 A0A1X7R6L9 A0A1J3JAS4 V7CSY1 M1UUR5 A0A165FND5 A0A165FP01
EC Number
5.2.1.8
3.4.-.-
3.4.-.-
Pubmed
EMBL
ODYU01001120
SOQ36888.1
NWSH01005009
PCG64498.1
AGBW02010257
OWR48855.1
+ More
KQ459580 KPI99192.1 BABH01000410 DQ311261 ABD36206.1 AGBW02011847 OWR46032.1 NWSH01001158 PCG72311.1 KQ460397 KPJ15308.1 RSAL01000023 RVE52282.1 KQ459439 KPJ01006.1 ODYU01004741 SOQ44939.1 ABLF02007382 ABLF02007383 ABLF02007384 ABLF02037453 ABLF02051027 KQ087230 KLT40675.1 CP028773 JQFK01000001 MQVM01000010 NHMM01000007 AWU74457.1 KGK40775.1 ONH74409.1 OUT20762.1 BDGI01000001 GAV26565.1 FN668690 CBK25231.2 MPUH01000693 OMJ75395.1 AMWG01000111 ELP32256.1 BX294148 CAD75841.1 AMCW01000067 EKK02182.1 AFAR01000210 EGF25797.1 GL832983 EGD78579.1 ANOF01000184 EMI23774.1 ANOQ01000104 EMI41953.1 GG692396 EER34520.1 ANMO01000177 EMB15364.1 KV784363 OEU13098.1 JXLN01010224 KPM05350.1 CP007031 AHF02631.1 AECK01000002 OBA17584.1 RHFJ01000033 RNE92675.1 DF237035 GAQ81654.1 HE650824 CCF58106.1 JH993041 EKX39262.1 LN609529 CEF66075.1 LSYU01000061 KXX64281.1 FR839628 CCA37613.1 BDGX01000021 GAV50365.1 CU928178 CAR28863.1 AZIL01000936 EWM25413.1 AFYH01246278 AFYH01246279 AFYH01246280 AFYH01246281 AAKM01000008 EDL44766.1 KQ235360 KMZ99969.1 KQ235038 KMZ93478.1 KQ234266 KMZ80912.1 LT615246 LT615263 LT635619 SCO66899.1 SCO72326.1 SGX77109.1 CP014586 ANZ76867.1 UFAJ01000077 SSD59008.1 KQ234811 KMZ87045.1 LECT01000038 KLU03342.1 GL378330 EFJ50807.1 KV454005 ODQ45149.1 KV453843 ODV88561.1 ANOH01000115 EMI57026.1 GG666545 EEN57283.1 LK052887 CDR38977.1 CAIF01000292 CCH46986.1 KV454215 ODQ56777.1 AAKM01000961 EDL42590.1 CP016250 ANQ09952.1 AHIQ01000260 EIF45632.1 LMYN01000004 KSA03885.1 MPUK01000006 ONH66795.1 LN679106 CEL62621.1 AAKM01000005 EDL45549.1 KQ235387 KMZ99692.1 KQ235046 KMZ93200.1 KQ234279 KMZ80403.1 LT615247 LT615264 LT635620 SCO67256.1 SCO72643.1 SGX77488.1 JH603170 EIC19580.1 MPUH01000239 OMJ85344.1 KZ454993 PKI82963.1 FXLY01000007 SMN21080.1 GEVM01016385 JAU89553.1 CM002288 ESW33224.1 AP006497 BAM81631.1 KV426077 KZV89273.1 KV426076 KZV89300.1
KQ459580 KPI99192.1 BABH01000410 DQ311261 ABD36206.1 AGBW02011847 OWR46032.1 NWSH01001158 PCG72311.1 KQ460397 KPJ15308.1 RSAL01000023 RVE52282.1 KQ459439 KPJ01006.1 ODYU01004741 SOQ44939.1 ABLF02007382 ABLF02007383 ABLF02007384 ABLF02037453 ABLF02051027 KQ087230 KLT40675.1 CP028773 JQFK01000001 MQVM01000010 NHMM01000007 AWU74457.1 KGK40775.1 ONH74409.1 OUT20762.1 BDGI01000001 GAV26565.1 FN668690 CBK25231.2 MPUH01000693 OMJ75395.1 AMWG01000111 ELP32256.1 BX294148 CAD75841.1 AMCW01000067 EKK02182.1 AFAR01000210 EGF25797.1 GL832983 EGD78579.1 ANOF01000184 EMI23774.1 ANOQ01000104 EMI41953.1 GG692396 EER34520.1 ANMO01000177 EMB15364.1 KV784363 OEU13098.1 JXLN01010224 KPM05350.1 CP007031 AHF02631.1 AECK01000002 OBA17584.1 RHFJ01000033 RNE92675.1 DF237035 GAQ81654.1 HE650824 CCF58106.1 JH993041 EKX39262.1 LN609529 CEF66075.1 LSYU01000061 KXX64281.1 FR839628 CCA37613.1 BDGX01000021 GAV50365.1 CU928178 CAR28863.1 AZIL01000936 EWM25413.1 AFYH01246278 AFYH01246279 AFYH01246280 AFYH01246281 AAKM01000008 EDL44766.1 KQ235360 KMZ99969.1 KQ235038 KMZ93478.1 KQ234266 KMZ80912.1 LT615246 LT615263 LT635619 SCO66899.1 SCO72326.1 SGX77109.1 CP014586 ANZ76867.1 UFAJ01000077 SSD59008.1 KQ234811 KMZ87045.1 LECT01000038 KLU03342.1 GL378330 EFJ50807.1 KV454005 ODQ45149.1 KV453843 ODV88561.1 ANOH01000115 EMI57026.1 GG666545 EEN57283.1 LK052887 CDR38977.1 CAIF01000292 CCH46986.1 KV454215 ODQ56777.1 AAKM01000961 EDL42590.1 CP016250 ANQ09952.1 AHIQ01000260 EIF45632.1 LMYN01000004 KSA03885.1 MPUK01000006 ONH66795.1 LN679106 CEL62621.1 AAKM01000005 EDL45549.1 KQ235387 KMZ99692.1 KQ235046 KMZ93200.1 KQ234279 KMZ80403.1 LT615247 LT615264 LT635620 SCO67256.1 SCO72643.1 SGX77488.1 JH603170 EIC19580.1 MPUH01000239 OMJ85344.1 KZ454993 PKI82963.1 FXLY01000007 SMN21080.1 GEVM01016385 JAU89553.1 CM002288 ESW33224.1 AP006497 BAM81631.1 KV426077 KZV89273.1 KV426076 KZV89300.1
Proteomes
UP000218220
UP000007151
UP000053268
UP000005204
UP000053240
UP000283053
+ More
UP000007819 UP000053611 UP000029867 UP000189274 UP000195871 UP000249293 UP000010959 UP000001025 UP000007993 UP000006222 UP000007799 UP000011996 UP000012028 UP000002037 UP000011529 UP000005275 UP000078553 UP000012960 UP000274052 UP000054558 UP000005220 UP000011087 UP000035682 UP000075766 UP000006853 UP000187013 UP000008536 UP000008672 UP000008333 UP000094565 UP000036367 UP000001058 UP000094455 UP000011885 UP000001554 UP000009328 UP000094112 UP000011116 UP000054251 UP000189513 UP000059188 UP000002964 UP000032180 UP000046392 UP000232875 UP000196158 UP000000226 UP000007014 UP000077266
UP000007819 UP000053611 UP000029867 UP000189274 UP000195871 UP000249293 UP000010959 UP000001025 UP000007993 UP000006222 UP000007799 UP000011996 UP000012028 UP000002037 UP000011529 UP000005275 UP000078553 UP000012960 UP000274052 UP000054558 UP000005220 UP000011087 UP000035682 UP000075766 UP000006853 UP000187013 UP000008536 UP000008672 UP000008333 UP000094565 UP000036367 UP000001058 UP000094455 UP000011885 UP000001554 UP000009328 UP000094112 UP000011116 UP000054251 UP000189513 UP000059188 UP000002964 UP000032180 UP000046392 UP000232875 UP000196158 UP000000226 UP000007014 UP000077266
Interpro
IPR002130
Cyclophilin-type_PPIase_dom
+ More
IPR029000 Cyclophilin-like_dom_sf
IPR015940 UBA
IPR009060 UBA-like_sf
IPR029488 Hmw/CFAP97D1
IPR024936 Cyclophilin-type_PPIase
IPR020892 Cyclophilin-type_PPIase_CS
IPR011993 PH-like_dom_sf
IPR000156 Ran_bind_dom
IPR008978 HSP20-like_chaperone
IPR007052 CS_dom
IPR039373 Peptidase_M28B
IPR007484 Peptidase_M28
IPR034308 VPS70
IPR029000 Cyclophilin-like_dom_sf
IPR015940 UBA
IPR009060 UBA-like_sf
IPR029488 Hmw/CFAP97D1
IPR024936 Cyclophilin-type_PPIase
IPR020892 Cyclophilin-type_PPIase_CS
IPR011993 PH-like_dom_sf
IPR000156 Ran_bind_dom
IPR008978 HSP20-like_chaperone
IPR007052 CS_dom
IPR039373 Peptidase_M28B
IPR007484 Peptidase_M28
IPR034308 VPS70
Gene 3D
ProteinModelPortal
A0A2H1V7R5
A0A2A4IYK9
A0A212F543
A0A194Q0T1
H9IV42
Q2F611
+ More
A0A212EX06 A0A2A4JK20 A0A194RBU3 A0A3S2NYK6 A0A194QC44 A0A2H1VVT0 J9KSD8 A0A0J0XHS7 A0A099P773 A0A1Q2YAJ4 D8MAZ2 A0A1R2BFH5 L7CGQ4 Q7UMR2 K5D633 F2AX20 F2UN80 M5SCB6 M5TDW4 C5M6A0 M2AZF2 A0A1E7F4I6 A0A132A4N0 W0E0J2 A0A1B7SM77 M0SV84 A0A3M8NRN9 A0A1Y1HSP4 H2AUQ6 L1IT53 A0A090LEW7 A0A162B4Q9 F2QQD5 A0A1Q3A3R1 C5DYK2 W7TPH9 H3A5C5 A5K7K7 A0A0J9TWG5 A0A0J9W0E8 A0A0J9SG55 A0A1G4HBM6 A0A1B2JFQ6 A0A376B2U8 A0A0J9SVE3 A0A0J1B9C4 D8TPQ4 A0A1E3NGA1 A0A1E4TA10 M0U4B7 M5UGQ2 C3YR41 A0A061AWC1 K0KUU1 A0A1E3NW22 A0A287I203 A5KDH9 A0A1B1E4J5 I2JRN0 A0A0V1Q606 A0A1V2L5F3 A0A0B7FXH5 A5K4H1 A0A0J9TXW6 A0A0J9TCI7 A0A0J9V388 A0A1G4HCM3 H8Z5U6 A0A0D9WU02 A0A0N5BJH8 A0A1R2C8K2 A0A2N1J8S6 A0A1X7R6L9 A0A1J3JAS4 V7CSY1 M1UUR5 A0A165FND5 A0A165FP01
A0A212EX06 A0A2A4JK20 A0A194RBU3 A0A3S2NYK6 A0A194QC44 A0A2H1VVT0 J9KSD8 A0A0J0XHS7 A0A099P773 A0A1Q2YAJ4 D8MAZ2 A0A1R2BFH5 L7CGQ4 Q7UMR2 K5D633 F2AX20 F2UN80 M5SCB6 M5TDW4 C5M6A0 M2AZF2 A0A1E7F4I6 A0A132A4N0 W0E0J2 A0A1B7SM77 M0SV84 A0A3M8NRN9 A0A1Y1HSP4 H2AUQ6 L1IT53 A0A090LEW7 A0A162B4Q9 F2QQD5 A0A1Q3A3R1 C5DYK2 W7TPH9 H3A5C5 A5K7K7 A0A0J9TWG5 A0A0J9W0E8 A0A0J9SG55 A0A1G4HBM6 A0A1B2JFQ6 A0A376B2U8 A0A0J9SVE3 A0A0J1B9C4 D8TPQ4 A0A1E3NGA1 A0A1E4TA10 M0U4B7 M5UGQ2 C3YR41 A0A061AWC1 K0KUU1 A0A1E3NW22 A0A287I203 A5KDH9 A0A1B1E4J5 I2JRN0 A0A0V1Q606 A0A1V2L5F3 A0A0B7FXH5 A5K4H1 A0A0J9TXW6 A0A0J9TCI7 A0A0J9V388 A0A1G4HCM3 H8Z5U6 A0A0D9WU02 A0A0N5BJH8 A0A1R2C8K2 A0A2N1J8S6 A0A1X7R6L9 A0A1J3JAS4 V7CSY1 M1UUR5 A0A165FND5 A0A165FP01
PDB
1QNG
E-value=2.84884e-07,
Score=124
Ontologies
GO
PANTHER
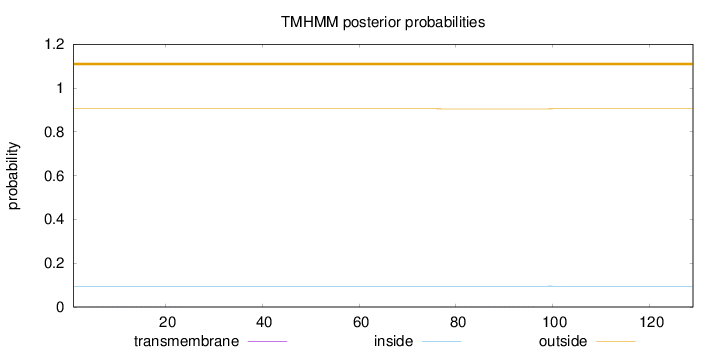
Topology
Length:
129
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00744
Exp number, first 60 AAs:
0.00357
Total prob of N-in:
0.09498
outside
1 - 129
Population Genetic Test Statistics
Pi
208.122377
Theta
52.141018
Tajima's D
1.231326
CLR
0.102001
CSRT
0.722613869306535
Interpretation
Uncertain
