Gene
KWMTBOMO14734
Pre Gene Modal
BGIBMGA014507
Annotation
vacuolar_ATPase?_subunit_M9.7_[Manduca_sexta]
Full name
V-type proton ATPase subunit
Location in the cell
PlasmaMembrane Reliability : 4.13
Sequence
CDS
ATGGGCTACTCCCTTATCCCGATCTTCGTTTTCTCCATCCTTTGGGGTGTGGTTGGTATTATTTGCCCCATCTTCGCACCTAAAGGACCAAACAGAGGGATTATTCAAGTGGTGTTAATATTAACGGCAGCGACTTGTTGGTTATTTTGGCTGTGTGCTTACATGGCACAGATGAACCCTCTCATCGGGCCCAGACTCAGCAATGAAACCCTAATCTGGATTTCACGCACTTGGGGAAACAAAATAAACAACACGCAAGCTTAA
Protein
MGYSLIPIFVFSILWGVVGIICPIFAPKGPNRGIIQVVLILTAATCWLFWLCAYMAQMNPLIGPRLSNETLIWISRTWGNKINNTQA
Summary
Description
Vacuolar ATPase is responsible for acidifying a variety of intracellular compartments in eukaryotic cells.
Subunit
Composed of at least 10 subunits.
Similarity
Belongs to the V-ATPase e1/e2 subunit family.
Feature
chain V-type proton ATPase subunit
Uniprot
O76959
S4NWG7
A0A2A4IZ97
I4DJ12
A0A212FB25
A0A2H1VR35
+ More
I4DN47 A0A2W1BHB9 A0A194R509 A0A0L7LC72 A0A1B0D3P9 U5EQM3 A0A1S3DLQ6 A0A1I8JUY2 A0A1L8DEG6 A0A084VH97 A0A2M3Z0M8 A0A2C9H8L1 A0A1I8JTR0 Q7QAP9 A0A182N4R6 A0A182QS91 Q1HRJ8 Q0IEF0 A0A2M4C3Z0 T1E2Q7 T1DRB1 A0A2M4AE41 A0A023EEI5 A0A2M4C3Y7 A0A2M4ACV5 A0A1Y9IVQ3 A0A2M4A6V4 A0A2M4AE73 A0A2M4C4U9 W5JJ96 A0A2M4CVF4 A0A0F7QZZ0 A0A2M4CVE3 A0A0L7QKJ5 D1FQ69 A0A067QXD0 J3JUH2 A0A1Q3FKU1 B0X1E3 A0A026WXW4 A0A2M4C4U3 A0A088AFI1 T1IC14 A0A0P4VWE5 G1K0S1 A0A2A3EDR7 V9IHU9 A0A0N0U7G8 A0A023F8Y7 A0A182Y7C6 A0A0V0G3W8 A0A182K8J9 A0A139WLV2 A0A158NVM9 A0A1W4X749 E0VJ42 A0A310S785 Q5MIS8 J9LV72 E2B2P9 A0A154PGW6 A0A232FB84 K7IN76 A0A195E463 A0A1B0GAE0 A0A1B0B9P9 A0A2M4B0B6 A0A151IMB7 A0A224Y280 A0A1D2NCM0 A0A146KKW7 E1ZWF8 A0A195FTQ0 R4WRA2 A0A0J7KU97 B4J1H4 F4WAG0 A0A0P6AZU9 E9GL66 A0A1J1J6B7 Q8SZ31 A0A0P5XZ98 B4LBQ3 B4H7X9 A0A1L8EFF6 A0A1W4URW7 B3NEA7 Q2VJI6 B4IAR7 B4QK61 Q9VP18 B4PDL2 Q4V4G1 A0A0L0CK79
I4DN47 A0A2W1BHB9 A0A194R509 A0A0L7LC72 A0A1B0D3P9 U5EQM3 A0A1S3DLQ6 A0A1I8JUY2 A0A1L8DEG6 A0A084VH97 A0A2M3Z0M8 A0A2C9H8L1 A0A1I8JTR0 Q7QAP9 A0A182N4R6 A0A182QS91 Q1HRJ8 Q0IEF0 A0A2M4C3Z0 T1E2Q7 T1DRB1 A0A2M4AE41 A0A023EEI5 A0A2M4C3Y7 A0A2M4ACV5 A0A1Y9IVQ3 A0A2M4A6V4 A0A2M4AE73 A0A2M4C4U9 W5JJ96 A0A2M4CVF4 A0A0F7QZZ0 A0A2M4CVE3 A0A0L7QKJ5 D1FQ69 A0A067QXD0 J3JUH2 A0A1Q3FKU1 B0X1E3 A0A026WXW4 A0A2M4C4U3 A0A088AFI1 T1IC14 A0A0P4VWE5 G1K0S1 A0A2A3EDR7 V9IHU9 A0A0N0U7G8 A0A023F8Y7 A0A182Y7C6 A0A0V0G3W8 A0A182K8J9 A0A139WLV2 A0A158NVM9 A0A1W4X749 E0VJ42 A0A310S785 Q5MIS8 J9LV72 E2B2P9 A0A154PGW6 A0A232FB84 K7IN76 A0A195E463 A0A1B0GAE0 A0A1B0B9P9 A0A2M4B0B6 A0A151IMB7 A0A224Y280 A0A1D2NCM0 A0A146KKW7 E1ZWF8 A0A195FTQ0 R4WRA2 A0A0J7KU97 B4J1H4 F4WAG0 A0A0P6AZU9 E9GL66 A0A1J1J6B7 Q8SZ31 A0A0P5XZ98 B4LBQ3 B4H7X9 A0A1L8EFF6 A0A1W4URW7 B3NEA7 Q2VJI6 B4IAR7 B4QK61 Q9VP18 B4PDL2 Q4V4G1 A0A0L0CK79
Pubmed
10358099
23622113
22651552
26354079
22118469
28756777
+ More
26227816 24438588 12364791 14747013 17210077 17204158 17510324 24330624 24945155 26483478 20920257 23761445 25937057 24845553 22516182 23537049 24508170 30249741 27129103 21736942 25474469 25244985 18362917 19820115 21347285 20566863 17244540 20798317 28648823 20075255 27289101 26823975 23691247 17994087 21719571 21292972 16085702 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 26108605
26227816 24438588 12364791 14747013 17210077 17204158 17510324 24330624 24945155 26483478 20920257 23761445 25937057 24845553 22516182 23537049 24508170 30249741 27129103 21736942 25474469 25244985 18362917 19820115 21347285 20566863 17244540 20798317 28648823 20075255 27289101 26823975 23691247 17994087 21719571 21292972 16085702 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 26108605
EMBL
AJ006029
CAA06822.1
GAIX01014665
JAA77895.1
NWSH01005009
PCG64500.1
+ More
AK401280 KQ459580 BAM17902.1 KPI99190.1 AGBW02009382 OWR50944.1 ODYU01003658 SOQ42704.1 AK402715 BAM19337.1 KZ150209 PZC72216.1 KQ460685 KPJ12767.1 JTDY01001716 KOB73093.1 AJVK01011005 GANO01003210 JAB56661.1 GFDF01009266 JAV04818.1 ATLV01013150 KE524842 KFB37341.1 GGFM01001322 MBW22073.1 APCN01005538 AAAB01008888 EAA08865.3 AXCN02000015 DQ440096 ABF18129.1 CH477695 EAT37149.1 EJY57885.1 GGFJ01010844 MBW59985.1 GALA01000474 JAA94378.1 GAMD01000705 JAB00886.1 GGFK01005557 MBW38878.1 JXUM01141252 JXUM01141253 GAPW01006533 KQ569212 JAC07065.1 KXJ68715.1 GGFJ01010915 MBW60056.1 GGFK01005117 MBW38438.1 GGFK01003160 MBW36481.1 GGFK01005766 MBW39087.1 GGFJ01011134 MBW60275.1 ADMH02001019 ETN64432.1 GGFL01005135 MBW69313.1 LC026146 BAR72391.1 GGFL01005134 MBW69312.1 KQ414940 KOC59147.1 EZ419969 ACZ28324.1 KK853137 KDR10794.1 APGK01016973 BT126886 KB739995 KB630343 KB631518 AEE61848.1 ENN82012.1 ERL83547.1 ERL84340.1 GFDL01006942 JAV28103.1 DS232258 EDS38592.1 KK107072 QOIP01000007 EZA60638.1 RLU20797.1 GGFJ01011189 MBW60330.1 ACPB03001840 GDKW01002676 JAI53919.1 JO495153 AEL79382.1 KZ288287 PBC29369.1 JR046707 AEY60237.1 KQ435710 KOX79728.1 GBBI01000805 JAC17907.1 GECL01003379 JAP02745.1 KQ971316 KYB28989.1 ADTU01027288 DS235219 EEB13398.1 KQ780658 OAD51929.1 AY826132 AAV90704.1 ABLF02013158 GL445210 EFN90031.1 KQ434902 KZC11105.1 NNAY01000480 OXU28091.1 KQ979701 KYN19644.1 CCAG010011415 JXJN01010515 GGFK01013165 MBW46486.1 KQ977085 KYN05906.1 GFTR01001673 JAW14753.1 LJIJ01000090 ODN02992.1 GDHC01021398 JAP97230.1 GL434829 EFN74481.1 KQ981281 KYN43269.1 AK417197 BAN20412.1 LBMM01003062 KMQ94012.1 CH916366 EDV95865.1 GL888048 EGI68843.1 GDIP01023001 LRGB01002190 JAM80714.1 KZS08703.1 GL732550 EFX79846.1 CVRI01000072 CRL07514.1 AY071161 AAL48783.1 GDIP01065273 JAM38442.1 CH940647 EDW68680.1 CH479219 EDW34769.1 GFDG01001381 JAV17418.1 CH954178 EDV52671.1 DQ079536 AAZ43020.1 CH480826 EDW44380.1 CM000363 CM002912 EDX11398.1 KMZ01039.1 AE014296 BT032649 AAF51743.2 ACD81663.1 CM000159 EDW95011.1 EDW95016.1 BT023045 AAY55461.2 JRES01000296 KNC32666.1
AK401280 KQ459580 BAM17902.1 KPI99190.1 AGBW02009382 OWR50944.1 ODYU01003658 SOQ42704.1 AK402715 BAM19337.1 KZ150209 PZC72216.1 KQ460685 KPJ12767.1 JTDY01001716 KOB73093.1 AJVK01011005 GANO01003210 JAB56661.1 GFDF01009266 JAV04818.1 ATLV01013150 KE524842 KFB37341.1 GGFM01001322 MBW22073.1 APCN01005538 AAAB01008888 EAA08865.3 AXCN02000015 DQ440096 ABF18129.1 CH477695 EAT37149.1 EJY57885.1 GGFJ01010844 MBW59985.1 GALA01000474 JAA94378.1 GAMD01000705 JAB00886.1 GGFK01005557 MBW38878.1 JXUM01141252 JXUM01141253 GAPW01006533 KQ569212 JAC07065.1 KXJ68715.1 GGFJ01010915 MBW60056.1 GGFK01005117 MBW38438.1 GGFK01003160 MBW36481.1 GGFK01005766 MBW39087.1 GGFJ01011134 MBW60275.1 ADMH02001019 ETN64432.1 GGFL01005135 MBW69313.1 LC026146 BAR72391.1 GGFL01005134 MBW69312.1 KQ414940 KOC59147.1 EZ419969 ACZ28324.1 KK853137 KDR10794.1 APGK01016973 BT126886 KB739995 KB630343 KB631518 AEE61848.1 ENN82012.1 ERL83547.1 ERL84340.1 GFDL01006942 JAV28103.1 DS232258 EDS38592.1 KK107072 QOIP01000007 EZA60638.1 RLU20797.1 GGFJ01011189 MBW60330.1 ACPB03001840 GDKW01002676 JAI53919.1 JO495153 AEL79382.1 KZ288287 PBC29369.1 JR046707 AEY60237.1 KQ435710 KOX79728.1 GBBI01000805 JAC17907.1 GECL01003379 JAP02745.1 KQ971316 KYB28989.1 ADTU01027288 DS235219 EEB13398.1 KQ780658 OAD51929.1 AY826132 AAV90704.1 ABLF02013158 GL445210 EFN90031.1 KQ434902 KZC11105.1 NNAY01000480 OXU28091.1 KQ979701 KYN19644.1 CCAG010011415 JXJN01010515 GGFK01013165 MBW46486.1 KQ977085 KYN05906.1 GFTR01001673 JAW14753.1 LJIJ01000090 ODN02992.1 GDHC01021398 JAP97230.1 GL434829 EFN74481.1 KQ981281 KYN43269.1 AK417197 BAN20412.1 LBMM01003062 KMQ94012.1 CH916366 EDV95865.1 GL888048 EGI68843.1 GDIP01023001 LRGB01002190 JAM80714.1 KZS08703.1 GL732550 EFX79846.1 CVRI01000072 CRL07514.1 AY071161 AAL48783.1 GDIP01065273 JAM38442.1 CH940647 EDW68680.1 CH479219 EDW34769.1 GFDG01001381 JAV17418.1 CH954178 EDV52671.1 DQ079536 AAZ43020.1 CH480826 EDW44380.1 CM000363 CM002912 EDX11398.1 KMZ01039.1 AE014296 BT032649 AAF51743.2 ACD81663.1 CM000159 EDW95011.1 EDW95016.1 BT023045 AAY55461.2 JRES01000296 KNC32666.1
Proteomes
UP000218220
UP000053268
UP000007151
UP000053240
UP000037510
UP000092462
+ More
UP000079169 UP000075900 UP000030765 UP000076407 UP000075840 UP000007062 UP000075884 UP000075886 UP000008820 UP000069940 UP000249989 UP000075920 UP000000673 UP000053825 UP000027135 UP000019118 UP000030742 UP000002320 UP000053097 UP000279307 UP000005203 UP000015103 UP000242457 UP000053105 UP000076408 UP000075881 UP000007266 UP000005205 UP000192223 UP000009046 UP000007819 UP000008237 UP000076502 UP000215335 UP000002358 UP000078492 UP000092444 UP000092460 UP000078542 UP000094527 UP000000311 UP000078541 UP000036403 UP000001070 UP000007755 UP000076858 UP000000305 UP000183832 UP000008792 UP000008744 UP000192221 UP000008711 UP000001292 UP000000304 UP000000803 UP000002282 UP000037069
UP000079169 UP000075900 UP000030765 UP000076407 UP000075840 UP000007062 UP000075884 UP000075886 UP000008820 UP000069940 UP000249989 UP000075920 UP000000673 UP000053825 UP000027135 UP000019118 UP000030742 UP000002320 UP000053097 UP000279307 UP000005203 UP000015103 UP000242457 UP000053105 UP000076408 UP000075881 UP000007266 UP000005205 UP000192223 UP000009046 UP000007819 UP000008237 UP000076502 UP000215335 UP000002358 UP000078492 UP000092444 UP000092460 UP000078542 UP000094527 UP000000311 UP000078541 UP000036403 UP000001070 UP000007755 UP000076858 UP000000305 UP000183832 UP000008792 UP000008744 UP000192221 UP000008711 UP000001292 UP000000304 UP000000803 UP000002282 UP000037069
Pfam
Interpro
Gene 3D
ProteinModelPortal
O76959
S4NWG7
A0A2A4IZ97
I4DJ12
A0A212FB25
A0A2H1VR35
+ More
I4DN47 A0A2W1BHB9 A0A194R509 A0A0L7LC72 A0A1B0D3P9 U5EQM3 A0A1S3DLQ6 A0A1I8JUY2 A0A1L8DEG6 A0A084VH97 A0A2M3Z0M8 A0A2C9H8L1 A0A1I8JTR0 Q7QAP9 A0A182N4R6 A0A182QS91 Q1HRJ8 Q0IEF0 A0A2M4C3Z0 T1E2Q7 T1DRB1 A0A2M4AE41 A0A023EEI5 A0A2M4C3Y7 A0A2M4ACV5 A0A1Y9IVQ3 A0A2M4A6V4 A0A2M4AE73 A0A2M4C4U9 W5JJ96 A0A2M4CVF4 A0A0F7QZZ0 A0A2M4CVE3 A0A0L7QKJ5 D1FQ69 A0A067QXD0 J3JUH2 A0A1Q3FKU1 B0X1E3 A0A026WXW4 A0A2M4C4U3 A0A088AFI1 T1IC14 A0A0P4VWE5 G1K0S1 A0A2A3EDR7 V9IHU9 A0A0N0U7G8 A0A023F8Y7 A0A182Y7C6 A0A0V0G3W8 A0A182K8J9 A0A139WLV2 A0A158NVM9 A0A1W4X749 E0VJ42 A0A310S785 Q5MIS8 J9LV72 E2B2P9 A0A154PGW6 A0A232FB84 K7IN76 A0A195E463 A0A1B0GAE0 A0A1B0B9P9 A0A2M4B0B6 A0A151IMB7 A0A224Y280 A0A1D2NCM0 A0A146KKW7 E1ZWF8 A0A195FTQ0 R4WRA2 A0A0J7KU97 B4J1H4 F4WAG0 A0A0P6AZU9 E9GL66 A0A1J1J6B7 Q8SZ31 A0A0P5XZ98 B4LBQ3 B4H7X9 A0A1L8EFF6 A0A1W4URW7 B3NEA7 Q2VJI6 B4IAR7 B4QK61 Q9VP18 B4PDL2 Q4V4G1 A0A0L0CK79
I4DN47 A0A2W1BHB9 A0A194R509 A0A0L7LC72 A0A1B0D3P9 U5EQM3 A0A1S3DLQ6 A0A1I8JUY2 A0A1L8DEG6 A0A084VH97 A0A2M3Z0M8 A0A2C9H8L1 A0A1I8JTR0 Q7QAP9 A0A182N4R6 A0A182QS91 Q1HRJ8 Q0IEF0 A0A2M4C3Z0 T1E2Q7 T1DRB1 A0A2M4AE41 A0A023EEI5 A0A2M4C3Y7 A0A2M4ACV5 A0A1Y9IVQ3 A0A2M4A6V4 A0A2M4AE73 A0A2M4C4U9 W5JJ96 A0A2M4CVF4 A0A0F7QZZ0 A0A2M4CVE3 A0A0L7QKJ5 D1FQ69 A0A067QXD0 J3JUH2 A0A1Q3FKU1 B0X1E3 A0A026WXW4 A0A2M4C4U3 A0A088AFI1 T1IC14 A0A0P4VWE5 G1K0S1 A0A2A3EDR7 V9IHU9 A0A0N0U7G8 A0A023F8Y7 A0A182Y7C6 A0A0V0G3W8 A0A182K8J9 A0A139WLV2 A0A158NVM9 A0A1W4X749 E0VJ42 A0A310S785 Q5MIS8 J9LV72 E2B2P9 A0A154PGW6 A0A232FB84 K7IN76 A0A195E463 A0A1B0GAE0 A0A1B0B9P9 A0A2M4B0B6 A0A151IMB7 A0A224Y280 A0A1D2NCM0 A0A146KKW7 E1ZWF8 A0A195FTQ0 R4WRA2 A0A0J7KU97 B4J1H4 F4WAG0 A0A0P6AZU9 E9GL66 A0A1J1J6B7 Q8SZ31 A0A0P5XZ98 B4LBQ3 B4H7X9 A0A1L8EFF6 A0A1W4URW7 B3NEA7 Q2VJI6 B4IAR7 B4QK61 Q9VP18 B4PDL2 Q4V4G1 A0A0L0CK79
Ontologies
PATHWAY
GO
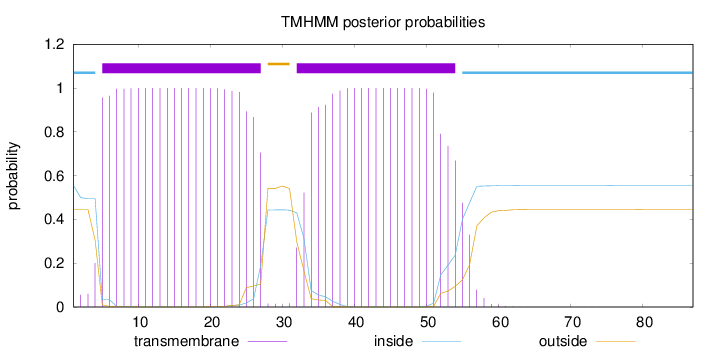
Topology
Subcellular location
Membrane
Length:
87
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
44.30754
Exp number, first 60 AAs:
44.28916
Total prob of N-in:
0.55459
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 27
outside
28 - 31
TMhelix
32 - 54
inside
55 - 87
Population Genetic Test Statistics
Pi
221.483274
Theta
179.269024
Tajima's D
0.715281
CLR
0.047288
CSRT
0.582020898955052
Interpretation
Uncertain
