Gene
KWMTBOMO14733
Annotation
hypothetical_protein_M513_10814?_partial_[Trichuris_suis]
Location in the cell
Nuclear Reliability : 2.197
Sequence
CDS
ATGGCCAGTCAAGAAATACAGGAACAACTGCAACAACTCTTGGCTGCAAATGCAGCGCTACAAAAACAACTTGAAGGAAATGAGAGAGAAACAAAAGTTTGTAGAGTTACAGTCAAATTGCCTACATTTTGGGCTGACAAACCAGCTGTTTGGTTTGCTCAAATTGAAGCTCAATTCCAAATTGCCAATATTGTGACAGATACTACTAAATATAATTATGTTGTAGGTCACCTTGACCAAAGAATAGCTGGAGAGGTTGAAGACATAATAACAAACCCACCAACTGAAGAAAAATATGAAAAATTAAAAAGCATACTTATTCAGCGCCTTTCTGTATCACAAGAAAAAAGAGTTCGACAACTTCTGAGTGATGAAGAACTTGGGGATAGAAAACCATCACAGTTCCTCCGACACCTACGCTCTCTTGCAGGAAGCACACTCACAGACGACAATATTATACGCCAACTTTGGTTACGCCGCTTGCCTCATCAGGCTCAAGCTATACTGGCAGCACAGGCTGACCTTGGCCTTGACAAATTGTCAGAACTAGCCGACAAAATTCTTGAGTTGGCCCAAACTCCTCCTATGGTATGTTCGACCGCAACATCTTCGAACAGTTTAACCCTTGGTTCATTAGCCAAACAACTAGAAGAGATCAGTAGCCAAGTTGCTGCCCTATCTAGGCGACAGAGCCGCCCTTATCGTGACAGATCATCATCTCGCGGCCCTAAAATTTGTTGATCACATTGA
Protein
MASQEIQEQLQQLLAANAALQKQLEGNERETKVCRVTVKLPTFWADKPAVWFAQIEAQFQIANIVTDTTKYNYVVGHLDQRIAGEVEDIITNPPTEEKYEKLKSILIQRLSVSQEKRVRQLLSDEELGDRKPSQFLRHLRSLAGSTLTDDNIIRQLWLRRLPHQAQAILAAQADLGLDKLSELADKILELAQTPPMVCSTATSSNSLTLGSLAKQLEEISSQVAALSRRQSRPYRDRSSSRGPKICXSH
Summary
Uniprot
A0A085LTM6
A0A085N4U2
A0A085NBV3
A0A0N5DV64
A0A2A4J8M7
A0A0J7KD10
+ More
A0A151IZ47 A0A151ITG2 A0A151J3D2 A0A2J7RGP3 A0A2J7PML8 A0A2J7QH56 A0A0J7K6I6 A0A2J7QJD3 A0A2J7RQK2 A0A0J7K7T5 E2BBH0 A0A2J7PCS9 A0A1W4WYX3 A0A0J7K112 W8B3F8 A0A2J7QTC8 A0A151JCH0 A0A1Y1MXV9 A0A0J7K6V3 A0A0L7QK36 A0A151K0A8 A0A0L7QJB5 A0A1W4XBP5 A0A2H1WXE7 A0A2J7Q198 A0A2A4K2H4 A0A2G8L3A3 A0A2G8LNJ3 A0A2J7PLR6 A0A1W4XIW9 A0A085MBU7 A0A1W4X6B7 A0A131YTY1 A0A0J7K6V1 T1IE13 A0A293MTK4 A0A2G8L664 A0A2J7Q5S3 W4Z1M4 A0A2J7PD24 A0A154NZG0 A0A026VRX8 A0A139W8G7 A0A2A4JHF4 D7GYF3 A0A2G8JGE7 A0A085NTM8 A0A293M349 A0A293M3N6 A0A085NBV0 A0A3L8D4U3 D7EJX5 W4YWZ1 A0A0N5DP72 A0A151JBY5 A0A085N631 A0A139WA14 A0A1S3CY78 A0A085M716 A0A154PMS5 W4ZJX1 A0A183NPV5 A0A095A584 A0A183JUP6 W4YUC1 A0A0J7NAK0 A0A183MCI1 C4QPF1 A0A139WPP8 A0A293N5Q9 A0A085MZ32 A0A182G9J3
A0A151IZ47 A0A151ITG2 A0A151J3D2 A0A2J7RGP3 A0A2J7PML8 A0A2J7QH56 A0A0J7K6I6 A0A2J7QJD3 A0A2J7RQK2 A0A0J7K7T5 E2BBH0 A0A2J7PCS9 A0A1W4WYX3 A0A0J7K112 W8B3F8 A0A2J7QTC8 A0A151JCH0 A0A1Y1MXV9 A0A0J7K6V3 A0A0L7QK36 A0A151K0A8 A0A0L7QJB5 A0A1W4XBP5 A0A2H1WXE7 A0A2J7Q198 A0A2A4K2H4 A0A2G8L3A3 A0A2G8LNJ3 A0A2J7PLR6 A0A1W4XIW9 A0A085MBU7 A0A1W4X6B7 A0A131YTY1 A0A0J7K6V1 T1IE13 A0A293MTK4 A0A2G8L664 A0A2J7Q5S3 W4Z1M4 A0A2J7PD24 A0A154NZG0 A0A026VRX8 A0A139W8G7 A0A2A4JHF4 D7GYF3 A0A2G8JGE7 A0A085NTM8 A0A293M349 A0A293M3N6 A0A085NBV0 A0A3L8D4U3 D7EJX5 W4YWZ1 A0A0N5DP72 A0A151JBY5 A0A085N631 A0A139WA14 A0A1S3CY78 A0A085M716 A0A154PMS5 W4ZJX1 A0A183NPV5 A0A095A584 A0A183JUP6 W4YUC1 A0A0J7NAK0 A0A183MCI1 C4QPF1 A0A139WPP8 A0A293N5Q9 A0A085MZ32 A0A182G9J3
Pubmed
EMBL
KL363297
KFD48322.1
KL367555
KFD64488.1
KL367519
KFD66949.1
+ More
NWSH01002598 PCG67884.1 LBMM01009142 KMQ88358.1 KQ980726 KYN14011.1 KQ981018 KYN10257.1 KQ980296 KYN16851.1 NEVH01003759 PNF40011.1 NEVH01023981 PNF17578.1 NEVH01014357 PNF27922.1 LBMM01013103 KMQ85816.1 NEVH01013558 PNF28672.1 NEVH01000707 PNF43124.1 LBMM01012075 KMQ86422.1 GL447057 EFN86960.1 NEVH01026435 PNF14140.1 LBMM01018126 KMQ83871.1 GAMC01010905 JAB95650.1 NEVH01011195 PNF31836.1 KQ979048 KYN23218.1 GEZM01017686 JAV90494.1 LBMM01012479 KMQ86203.1 KQ414987 KOC58988.1 KQ981301 KYN43041.1 LHQN01021927 LHQN01025379 KOC58628.1 KOC58659.1 ODYU01011787 SOQ57731.1 NEVH01019603 PNF22346.1 NWSH01000267 PCG77880.1 MRZV01001127 MRZV01000236 PIK40395.1 PIK54731.1 MRZV01000025 PIK61772.1 NEVH01024424 PNF17276.1 KL363205 KFD54693.1 GEDV01006140 JAP82417.1 LBMM01012923 LBMM01012917 KMQ85926.1 KMQ85930.1 ACPB03037918 GFWV01018540 MAA43268.1 MRZV01000203 PIK55728.1 NEVH01017547 PNF23930.1 AAGJ04018350 NEVH01026394 PNF14234.1 KQ434784 KZC04993.1 KK111495 EZA46390.1 KQ973343 KXZ75568.1 NWSH01001575 PCG70822.1 GG695519 EFA13383.1 MRZV01002078 PIK34788.1 KL367475 KFD72824.1 GFWV01009910 MAA34639.1 GFWV01009909 MAA34638.1 KFD66946.1 QOIP01000013 RLU15256.1 KQ971759 EFA12902.2 AAGJ04150373 KQ979103 KYN22624.1 KL367547 KFD64927.1 KQ971745 KYB24749.1 KL363221 KFD53012.1 KQ434992 KZC13185.1 AAGJ04133382 UZAL01010187 VDP03243.1 KL252356 KGB42042.1 UZAK01014297 VDP04051.1 AAGJ04172791 LBMM01007548 KMQ89630.1 UZAI01011057 VDP08268.1 CABG01000134 CAZ37338.2 KQ971307 KYB29887.1 GFWV01023686 MAA48413.1 KL367594 KFD62478.1 JXUM01049417 KQ561603 KXJ78027.1
NWSH01002598 PCG67884.1 LBMM01009142 KMQ88358.1 KQ980726 KYN14011.1 KQ981018 KYN10257.1 KQ980296 KYN16851.1 NEVH01003759 PNF40011.1 NEVH01023981 PNF17578.1 NEVH01014357 PNF27922.1 LBMM01013103 KMQ85816.1 NEVH01013558 PNF28672.1 NEVH01000707 PNF43124.1 LBMM01012075 KMQ86422.1 GL447057 EFN86960.1 NEVH01026435 PNF14140.1 LBMM01018126 KMQ83871.1 GAMC01010905 JAB95650.1 NEVH01011195 PNF31836.1 KQ979048 KYN23218.1 GEZM01017686 JAV90494.1 LBMM01012479 KMQ86203.1 KQ414987 KOC58988.1 KQ981301 KYN43041.1 LHQN01021927 LHQN01025379 KOC58628.1 KOC58659.1 ODYU01011787 SOQ57731.1 NEVH01019603 PNF22346.1 NWSH01000267 PCG77880.1 MRZV01001127 MRZV01000236 PIK40395.1 PIK54731.1 MRZV01000025 PIK61772.1 NEVH01024424 PNF17276.1 KL363205 KFD54693.1 GEDV01006140 JAP82417.1 LBMM01012923 LBMM01012917 KMQ85926.1 KMQ85930.1 ACPB03037918 GFWV01018540 MAA43268.1 MRZV01000203 PIK55728.1 NEVH01017547 PNF23930.1 AAGJ04018350 NEVH01026394 PNF14234.1 KQ434784 KZC04993.1 KK111495 EZA46390.1 KQ973343 KXZ75568.1 NWSH01001575 PCG70822.1 GG695519 EFA13383.1 MRZV01002078 PIK34788.1 KL367475 KFD72824.1 GFWV01009910 MAA34639.1 GFWV01009909 MAA34638.1 KFD66946.1 QOIP01000013 RLU15256.1 KQ971759 EFA12902.2 AAGJ04150373 KQ979103 KYN22624.1 KL367547 KFD64927.1 KQ971745 KYB24749.1 KL363221 KFD53012.1 KQ434992 KZC13185.1 AAGJ04133382 UZAL01010187 VDP03243.1 KL252356 KGB42042.1 UZAK01014297 VDP04051.1 AAGJ04172791 LBMM01007548 KMQ89630.1 UZAI01011057 VDP08268.1 CABG01000134 CAZ37338.2 KQ971307 KYB29887.1 GFWV01023686 MAA48413.1 KL367594 KFD62478.1 JXUM01049417 KQ561603 KXJ78027.1
Proteomes
Pfam
Interpro
IPR021109
Peptidase_aspartic_dom_sf
+ More
IPR001969 Aspartic_peptidase_AS
IPR000477 RT_dom
IPR001995 Peptidase_A2_cat
IPR034132 RP_Saci-like
IPR041588 Integrase_H2C2
IPR041577 RT_RNaseH_2
IPR041373 RT_RNaseH
IPR018289 MULE_transposase_dom
IPR036397 RNaseH_sf
IPR001584 Integrase_cat-core
IPR012337 RNaseH-like_sf
IPR001969 Aspartic_peptidase_AS
IPR000477 RT_dom
IPR001995 Peptidase_A2_cat
IPR034132 RP_Saci-like
IPR041588 Integrase_H2C2
IPR041577 RT_RNaseH_2
IPR041373 RT_RNaseH
IPR018289 MULE_transposase_dom
IPR036397 RNaseH_sf
IPR001584 Integrase_cat-core
IPR012337 RNaseH-like_sf
Gene 3D
ProteinModelPortal
A0A085LTM6
A0A085N4U2
A0A085NBV3
A0A0N5DV64
A0A2A4J8M7
A0A0J7KD10
+ More
A0A151IZ47 A0A151ITG2 A0A151J3D2 A0A2J7RGP3 A0A2J7PML8 A0A2J7QH56 A0A0J7K6I6 A0A2J7QJD3 A0A2J7RQK2 A0A0J7K7T5 E2BBH0 A0A2J7PCS9 A0A1W4WYX3 A0A0J7K112 W8B3F8 A0A2J7QTC8 A0A151JCH0 A0A1Y1MXV9 A0A0J7K6V3 A0A0L7QK36 A0A151K0A8 A0A0L7QJB5 A0A1W4XBP5 A0A2H1WXE7 A0A2J7Q198 A0A2A4K2H4 A0A2G8L3A3 A0A2G8LNJ3 A0A2J7PLR6 A0A1W4XIW9 A0A085MBU7 A0A1W4X6B7 A0A131YTY1 A0A0J7K6V1 T1IE13 A0A293MTK4 A0A2G8L664 A0A2J7Q5S3 W4Z1M4 A0A2J7PD24 A0A154NZG0 A0A026VRX8 A0A139W8G7 A0A2A4JHF4 D7GYF3 A0A2G8JGE7 A0A085NTM8 A0A293M349 A0A293M3N6 A0A085NBV0 A0A3L8D4U3 D7EJX5 W4YWZ1 A0A0N5DP72 A0A151JBY5 A0A085N631 A0A139WA14 A0A1S3CY78 A0A085M716 A0A154PMS5 W4ZJX1 A0A183NPV5 A0A095A584 A0A183JUP6 W4YUC1 A0A0J7NAK0 A0A183MCI1 C4QPF1 A0A139WPP8 A0A293N5Q9 A0A085MZ32 A0A182G9J3
A0A151IZ47 A0A151ITG2 A0A151J3D2 A0A2J7RGP3 A0A2J7PML8 A0A2J7QH56 A0A0J7K6I6 A0A2J7QJD3 A0A2J7RQK2 A0A0J7K7T5 E2BBH0 A0A2J7PCS9 A0A1W4WYX3 A0A0J7K112 W8B3F8 A0A2J7QTC8 A0A151JCH0 A0A1Y1MXV9 A0A0J7K6V3 A0A0L7QK36 A0A151K0A8 A0A0L7QJB5 A0A1W4XBP5 A0A2H1WXE7 A0A2J7Q198 A0A2A4K2H4 A0A2G8L3A3 A0A2G8LNJ3 A0A2J7PLR6 A0A1W4XIW9 A0A085MBU7 A0A1W4X6B7 A0A131YTY1 A0A0J7K6V1 T1IE13 A0A293MTK4 A0A2G8L664 A0A2J7Q5S3 W4Z1M4 A0A2J7PD24 A0A154NZG0 A0A026VRX8 A0A139W8G7 A0A2A4JHF4 D7GYF3 A0A2G8JGE7 A0A085NTM8 A0A293M349 A0A293M3N6 A0A085NBV0 A0A3L8D4U3 D7EJX5 W4YWZ1 A0A0N5DP72 A0A151JBY5 A0A085N631 A0A139WA14 A0A1S3CY78 A0A085M716 A0A154PMS5 W4ZJX1 A0A183NPV5 A0A095A584 A0A183JUP6 W4YUC1 A0A0J7NAK0 A0A183MCI1 C4QPF1 A0A139WPP8 A0A293N5Q9 A0A085MZ32 A0A182G9J3
Ontologies
KEGG
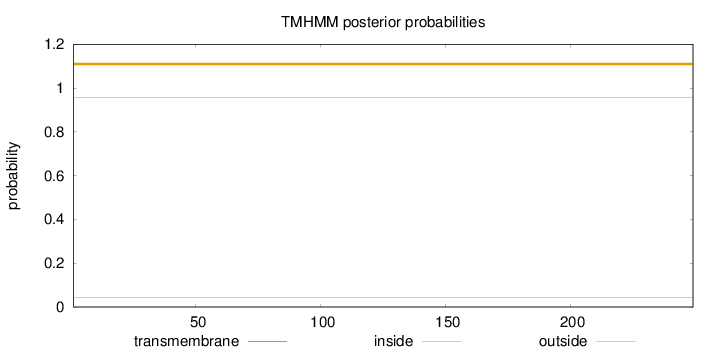
Topology
Length:
249
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000830000000000001
Exp number, first 60 AAs:
0.00049
Total prob of N-in:
0.04484
outside
1 - 249
Population Genetic Test Statistics
Pi
189.816637
Theta
19.798322
Tajima's D
1.899191
CLR
37.043455
CSRT
0.869606519674016
Interpretation
Uncertain
