Gene
KWMTBOMO14731
Pre Gene Modal
BGIBMGA003828
Annotation
Transcription_initiation_factor_TFIID_subunit_10_[Operophtera_brumata]
Full name
Transcription initiation factor TFIID subunit 10
Alternative Name
Transcription initiation factor TFIID 30 kDa subunit
Location in the cell
Extracellular Reliability : 1.59 Nuclear Reliability : 2.246
Sequence
CDS
ATGAGCCGTATCGGTTCACCATCGGTCGGTATGGACGACGACGGTTGCGCAGGACACGCTTTAGGCGACTTCCTTCTTCAGCTTGAGAATTATAATCCATCCATACCCGATTCCGTGGTCGCATATTATTTGAATTTAACCGGATTCGAGTCACAGGATCCGAGACTTGTCAGATTGATCGCATTGGCATCACAGAAATTCTTATCTGACATTGTAAATGATGCACTGCAGCACTGCAAGATGCGTACGTCGAATCAAATAAATCAAAGCTCCAAAAACCAGAAGGGACCCAAAGAGAAGAAATATGTCATGACCATGGACGATTTGGCTCCCGCGTTACAAGAGTATGGCATAACAGCAAAGAAACCGCACTATTTTGTTTGA
Protein
MSRIGSPSVGMDDDGCAGHALGDFLLQLENYNPSIPDSVVAYYLNLTGFESQDPRLVRLIALASQKFLSDIVNDALQHCKMRTSNQINQSSKNQKGPKEKKYVMTMDDLAPALQEYGITAKKPHYFV
Summary
Description
TFIID is a multimeric protein complex that plays a central role in mediating promoter responses to various activators and repressors.
TAFs are components of the transcription factor IID (TFIID) complex, PCAF histone acetylase complex and TBP-free TAFII complex (TFTC). TIIFD is a multimeric protein complex that plays a central role in mediating promoter responses to various activators and repressors. May regulate cyclin E expression.
TAFs are components of the transcription factor IID (TFIID) complex, PCAF histone acetylase complex and TBP-free TAFII complex (TFTC). TIIFD is a multimeric protein complex that plays a central role in mediating promoter responses to various activators and repressors. May regulate cyclin E expression.
Subunit
TFIID and PCAF are composed of TATA binding protein (TBP) and a number of TBP-associated factors (TAFs). TBP is not part of TFTC. Component of the PCAF complex, at least composed of TADA2L/ADA2, TADA3L/ADA3, SUPT3H, TAF5L, TAF6L, TAF9, TAF10, TAF12 and TRRAP. Component of the TFTC-HAT complex, at least composed of TAF5L, TAF6L, TADA3L, SUPT3H, TAF2, TAF4, TAF5, GCN5L2/GCN5, TAF10 and TRRAP. Component of the STAGA transcription coactivator-HAT complex, at least composed of SUPT3H, GCN5L2, TAF5L, TAF6L, SUPT7L, TADA3L, TAD1L, TAF10, TAF12, TRRAP and TAF9. The STAGA core complex is associated with a subcomplex required for histone deubiquitination composed of ATXN7L3, ENY2 and USP22 (By similarity). Interacts with TAF3 (PubMed:11438666). Interacts with LOXL2 (By similarity).
Similarity
Belongs to the TAF10 family.
Keywords
Acetylation
Complete proteome
Methylation
Nucleus
Phosphoprotein
Reference proteome
Transcription
Transcription regulation
Feature
chain Transcription initiation factor TFIID subunit 10
Uniprot
H9J2U1
A0A2W1BET2
A0A2A4J0N3
A0A0L7KEB0
S4P4X4
A0A2H1WBD8
+ More
A0A1E1W6M4 A0A194Q1I7 A0A194R509 A0A2J7PYT2 A0A0K8TPT8 C3XPP3 A0A1B0DH81 A0A1Q3FG34 J3JZ71 B0WUQ2 A0A182P4B1 R4ULY9 A0A182SKC6 A0A023EFC4 A0A067RI97 A0A1I8PU81 A0A182RK78 W5JT55 Q178Y1 A0A182QJ97 A0A084VFE4 A0A182NTD3 A0A2T7PPV3 A0A2B4RQR4 A0A182J3M2 A0A182WFJ1 A0A182V0V2 A0A182U5A3 Q7Q8I7 A0A182F0T6 A0A3N0YT46 A0A2M3Z4H5 Q1LVD6 Q1LVD5 M9VAC9 U5EQC9 A0A2M4C2X1 A0A182X5Y8 A0A182YQI3 A0A182KN82 A0A1A8DTE1 A0A1A8UYN2 A0A1A8QV20 A0A1A8JZU8 C1BLB8 A0A0P7WH99 A0A1A7XWE6 A0A1W4XXH4 A0A0L8FHD8 A0A0E9WVK1 C1BH11 W5M3F8 A0A1A8EX70 C1BIA8 B5X8E6 H3D363 Q8K0H5 A0A3S3P410 A0A1L8DGY8 A0A1B6D4A5 A0A060YFI9 H2UQS7 A0A1A8L6I8 F7FRQ9 A0A3B1JZM9 A0A093F905 A0A1A6HQN7 G7NE05 G1SRB2 A0A093KGM3 A0A091U720 A0A087VEB8 A0A091LHN5 A0A091JJP1 A0A341CE20 A0A091V8A0 R7V2A5 A0A091TRJ8 A0A094KSU6 A0A1B0FR10 A0A151MYK0 G3U484 A0A093Q2L5 A0A2M4AM13 C1BY99 B5X692 A0A3P8XLE2 A0A091LSB3 D2HR58 V4BRI6 C1BEY2 B4F7B2 A0A091D2J5
A0A1E1W6M4 A0A194Q1I7 A0A194R509 A0A2J7PYT2 A0A0K8TPT8 C3XPP3 A0A1B0DH81 A0A1Q3FG34 J3JZ71 B0WUQ2 A0A182P4B1 R4ULY9 A0A182SKC6 A0A023EFC4 A0A067RI97 A0A1I8PU81 A0A182RK78 W5JT55 Q178Y1 A0A182QJ97 A0A084VFE4 A0A182NTD3 A0A2T7PPV3 A0A2B4RQR4 A0A182J3M2 A0A182WFJ1 A0A182V0V2 A0A182U5A3 Q7Q8I7 A0A182F0T6 A0A3N0YT46 A0A2M3Z4H5 Q1LVD6 Q1LVD5 M9VAC9 U5EQC9 A0A2M4C2X1 A0A182X5Y8 A0A182YQI3 A0A182KN82 A0A1A8DTE1 A0A1A8UYN2 A0A1A8QV20 A0A1A8JZU8 C1BLB8 A0A0P7WH99 A0A1A7XWE6 A0A1W4XXH4 A0A0L8FHD8 A0A0E9WVK1 C1BH11 W5M3F8 A0A1A8EX70 C1BIA8 B5X8E6 H3D363 Q8K0H5 A0A3S3P410 A0A1L8DGY8 A0A1B6D4A5 A0A060YFI9 H2UQS7 A0A1A8L6I8 F7FRQ9 A0A3B1JZM9 A0A093F905 A0A1A6HQN7 G7NE05 G1SRB2 A0A093KGM3 A0A091U720 A0A087VEB8 A0A091LHN5 A0A091JJP1 A0A341CE20 A0A091V8A0 R7V2A5 A0A091TRJ8 A0A094KSU6 A0A1B0FR10 A0A151MYK0 G3U484 A0A093Q2L5 A0A2M4AM13 C1BY99 B5X692 A0A3P8XLE2 A0A091LSB3 D2HR58 V4BRI6 C1BEY2 B4F7B2 A0A091D2J5
Pubmed
19121390
28756777
26227816
23622113
26354079
26369729
+ More
18563158 22516182 24945155 26483478 24845553 20920257 23761445 17510324 24438588 12364791 14747013 17210077 23594743 25244985 20966253 25613341 24755649 20433749 15496914 10469660 16141072 15489334 11438666 21183079 25959397 21551351 17495919 25329095 22002653 21993624 23254933 22293439 25069045 20010809 15057822 15632090
18563158 22516182 24945155 26483478 24845553 20920257 23761445 17510324 24438588 12364791 14747013 17210077 23594743 25244985 20966253 25613341 24755649 20433749 15496914 10469660 16141072 15489334 11438666 21183079 25959397 21551351 17495919 25329095 22002653 21993624 23254933 22293439 25069045 20010809 15057822 15632090
EMBL
BABH01036846
KZ150209
PZC72214.1
NWSH01004536
PCG64972.1
JTDY01010031
+ More
KOB61647.1 GAIX01011005 JAA81555.1 ODYU01007535 SOQ50391.1 GDQN01008447 GDQN01008375 JAT82607.1 JAT82679.1 KQ459580 KPI99188.1 KQ460685 KPJ12767.1 NEVH01020342 PNF21507.1 GDAI01001460 JAI16143.1 GG666451 EEN69914.1 AJVK01060754 GFDL01008531 JAV26514.1 BT128552 AEE63509.1 DS232108 EDS35038.1 KC632286 AGM32100.1 JXUM01085952 GAPW01005591 GAPW01005590 KQ563535 JAC08007.1 KXJ73700.1 KK852496 KDR22733.1 ADMH02000444 ETN66463.1 CH477357 EAT42761.1 AXCN02000713 ATLV01012384 KE524787 KFB36688.1 PZQS01000002 PVD35452.1 LSMT01000337 PFX19951.1 AAAB01008944 EAA10055.4 RJVU01027559 ROL49180.1 GGFM01002663 MBW23414.1 BX784025 BC133967 AAI33968.1 KC161218 AGJ76546.1 GANO01004309 JAB55562.1 GGFJ01010512 MBW59653.1 HADZ01010919 HAEA01008757 SBQ37237.1 HADY01008418 HAEJ01012728 SBS53185.1 HAEH01005124 HAEI01006277 SBR96879.1 HAED01018681 HAEE01005486 SBR25506.1 BT075397 ACO09821.1 JARO02008277 KPP62928.1 HADW01018001 HADX01000225 SBP22457.1 KQ431609 KOF63085.1 GBXM01014148 JAH94429.1 BT073890 ACO08314.1 AHAT01009638 HAEB01003917 HAEC01008597 SBQ50444.1 BT074337 FR904730 ACO08761.1 CDQ70660.1 BT047315 ACI67116.1 AK047115 AJ249987 BC031418 BC080311 NCKU01004553 NCKU01003389 NCKU01003365 RWS05779.1 RWS07612.1 RWS07664.1 GFDF01008366 JAV05718.1 GEDC01019107 GEDC01018820 GEDC01016767 GEDC01008178 GEDC01004600 GEDC01000355 JAS18191.1 JAS18478.1 JAS20531.1 JAS29120.1 JAS32698.1 JAS36943.1 FR908228 CDQ87910.1 HAEF01002614 HAEG01004191 SBR39996.1 KK619954 KFV50669.1 LZPO01017338 OBS80571.1 CM001266 EHH23133.1 AAGW02008015 KK585888 KFV96980.1 KK419672 KFQ86256.1 KL490132 KFO10960.1 KL320753 KFP54823.1 KK502044 KFP19965.1 KL410773 KFQ99531.1 AMQN01005350 KB295796 ELU12612.1 KK464421 KFQ80984.1 KL267029 KFZ62308.1 CCAG010010711 AKHW03004616 KYO29617.1 KL415783 KFW80650.1 GGFK01008499 MBW41820.1 BT079578 ACO14002.1 BT046561 BT047939 BT125391 ACI66362.1 ACI67740.1 ADM16132.1 KK509003 KFP62398.1 GL193208 EFB14163.1 KB202283 ESO91494.1 BT073161 ACO07585.1 AABR07071958 BC168203 CH473956 AAI68203.1 EDM18002.1 KN123504 KFO24718.1
KOB61647.1 GAIX01011005 JAA81555.1 ODYU01007535 SOQ50391.1 GDQN01008447 GDQN01008375 JAT82607.1 JAT82679.1 KQ459580 KPI99188.1 KQ460685 KPJ12767.1 NEVH01020342 PNF21507.1 GDAI01001460 JAI16143.1 GG666451 EEN69914.1 AJVK01060754 GFDL01008531 JAV26514.1 BT128552 AEE63509.1 DS232108 EDS35038.1 KC632286 AGM32100.1 JXUM01085952 GAPW01005591 GAPW01005590 KQ563535 JAC08007.1 KXJ73700.1 KK852496 KDR22733.1 ADMH02000444 ETN66463.1 CH477357 EAT42761.1 AXCN02000713 ATLV01012384 KE524787 KFB36688.1 PZQS01000002 PVD35452.1 LSMT01000337 PFX19951.1 AAAB01008944 EAA10055.4 RJVU01027559 ROL49180.1 GGFM01002663 MBW23414.1 BX784025 BC133967 AAI33968.1 KC161218 AGJ76546.1 GANO01004309 JAB55562.1 GGFJ01010512 MBW59653.1 HADZ01010919 HAEA01008757 SBQ37237.1 HADY01008418 HAEJ01012728 SBS53185.1 HAEH01005124 HAEI01006277 SBR96879.1 HAED01018681 HAEE01005486 SBR25506.1 BT075397 ACO09821.1 JARO02008277 KPP62928.1 HADW01018001 HADX01000225 SBP22457.1 KQ431609 KOF63085.1 GBXM01014148 JAH94429.1 BT073890 ACO08314.1 AHAT01009638 HAEB01003917 HAEC01008597 SBQ50444.1 BT074337 FR904730 ACO08761.1 CDQ70660.1 BT047315 ACI67116.1 AK047115 AJ249987 BC031418 BC080311 NCKU01004553 NCKU01003389 NCKU01003365 RWS05779.1 RWS07612.1 RWS07664.1 GFDF01008366 JAV05718.1 GEDC01019107 GEDC01018820 GEDC01016767 GEDC01008178 GEDC01004600 GEDC01000355 JAS18191.1 JAS18478.1 JAS20531.1 JAS29120.1 JAS32698.1 JAS36943.1 FR908228 CDQ87910.1 HAEF01002614 HAEG01004191 SBR39996.1 KK619954 KFV50669.1 LZPO01017338 OBS80571.1 CM001266 EHH23133.1 AAGW02008015 KK585888 KFV96980.1 KK419672 KFQ86256.1 KL490132 KFO10960.1 KL320753 KFP54823.1 KK502044 KFP19965.1 KL410773 KFQ99531.1 AMQN01005350 KB295796 ELU12612.1 KK464421 KFQ80984.1 KL267029 KFZ62308.1 CCAG010010711 AKHW03004616 KYO29617.1 KL415783 KFW80650.1 GGFK01008499 MBW41820.1 BT079578 ACO14002.1 BT046561 BT047939 BT125391 ACI66362.1 ACI67740.1 ADM16132.1 KK509003 KFP62398.1 GL193208 EFB14163.1 KB202283 ESO91494.1 BT073161 ACO07585.1 AABR07071958 BC168203 CH473956 AAI68203.1 EDM18002.1 KN123504 KFO24718.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053268
UP000053240
UP000235965
+ More
UP000001554 UP000092462 UP000002320 UP000075885 UP000075901 UP000069940 UP000249989 UP000027135 UP000095300 UP000075900 UP000000673 UP000008820 UP000075886 UP000030765 UP000075884 UP000245119 UP000225706 UP000075880 UP000075920 UP000075903 UP000075902 UP000007062 UP000069272 UP000000437 UP000076407 UP000076408 UP000075882 UP000034805 UP000192224 UP000053454 UP000018468 UP000193380 UP000087266 UP000007303 UP000000589 UP000285301 UP000005226 UP000002280 UP000018467 UP000092124 UP000001811 UP000053119 UP000252040 UP000053283 UP000014760 UP000092444 UP000050525 UP000007646 UP000265140 UP000030746 UP000002494 UP000028990
UP000001554 UP000092462 UP000002320 UP000075885 UP000075901 UP000069940 UP000249989 UP000027135 UP000095300 UP000075900 UP000000673 UP000008820 UP000075886 UP000030765 UP000075884 UP000245119 UP000225706 UP000075880 UP000075920 UP000075903 UP000075902 UP000007062 UP000069272 UP000000437 UP000076407 UP000076408 UP000075882 UP000034805 UP000192224 UP000053454 UP000018468 UP000193380 UP000087266 UP000007303 UP000000589 UP000285301 UP000005226 UP000002280 UP000018467 UP000092124 UP000001811 UP000053119 UP000252040 UP000053283 UP000014760 UP000092444 UP000050525 UP000007646 UP000265140 UP000030746 UP000002494 UP000028990
Gene 3D
CDD
ProteinModelPortal
H9J2U1
A0A2W1BET2
A0A2A4J0N3
A0A0L7KEB0
S4P4X4
A0A2H1WBD8
+ More
A0A1E1W6M4 A0A194Q1I7 A0A194R509 A0A2J7PYT2 A0A0K8TPT8 C3XPP3 A0A1B0DH81 A0A1Q3FG34 J3JZ71 B0WUQ2 A0A182P4B1 R4ULY9 A0A182SKC6 A0A023EFC4 A0A067RI97 A0A1I8PU81 A0A182RK78 W5JT55 Q178Y1 A0A182QJ97 A0A084VFE4 A0A182NTD3 A0A2T7PPV3 A0A2B4RQR4 A0A182J3M2 A0A182WFJ1 A0A182V0V2 A0A182U5A3 Q7Q8I7 A0A182F0T6 A0A3N0YT46 A0A2M3Z4H5 Q1LVD6 Q1LVD5 M9VAC9 U5EQC9 A0A2M4C2X1 A0A182X5Y8 A0A182YQI3 A0A182KN82 A0A1A8DTE1 A0A1A8UYN2 A0A1A8QV20 A0A1A8JZU8 C1BLB8 A0A0P7WH99 A0A1A7XWE6 A0A1W4XXH4 A0A0L8FHD8 A0A0E9WVK1 C1BH11 W5M3F8 A0A1A8EX70 C1BIA8 B5X8E6 H3D363 Q8K0H5 A0A3S3P410 A0A1L8DGY8 A0A1B6D4A5 A0A060YFI9 H2UQS7 A0A1A8L6I8 F7FRQ9 A0A3B1JZM9 A0A093F905 A0A1A6HQN7 G7NE05 G1SRB2 A0A093KGM3 A0A091U720 A0A087VEB8 A0A091LHN5 A0A091JJP1 A0A341CE20 A0A091V8A0 R7V2A5 A0A091TRJ8 A0A094KSU6 A0A1B0FR10 A0A151MYK0 G3U484 A0A093Q2L5 A0A2M4AM13 C1BY99 B5X692 A0A3P8XLE2 A0A091LSB3 D2HR58 V4BRI6 C1BEY2 B4F7B2 A0A091D2J5
A0A1E1W6M4 A0A194Q1I7 A0A194R509 A0A2J7PYT2 A0A0K8TPT8 C3XPP3 A0A1B0DH81 A0A1Q3FG34 J3JZ71 B0WUQ2 A0A182P4B1 R4ULY9 A0A182SKC6 A0A023EFC4 A0A067RI97 A0A1I8PU81 A0A182RK78 W5JT55 Q178Y1 A0A182QJ97 A0A084VFE4 A0A182NTD3 A0A2T7PPV3 A0A2B4RQR4 A0A182J3M2 A0A182WFJ1 A0A182V0V2 A0A182U5A3 Q7Q8I7 A0A182F0T6 A0A3N0YT46 A0A2M3Z4H5 Q1LVD6 Q1LVD5 M9VAC9 U5EQC9 A0A2M4C2X1 A0A182X5Y8 A0A182YQI3 A0A182KN82 A0A1A8DTE1 A0A1A8UYN2 A0A1A8QV20 A0A1A8JZU8 C1BLB8 A0A0P7WH99 A0A1A7XWE6 A0A1W4XXH4 A0A0L8FHD8 A0A0E9WVK1 C1BH11 W5M3F8 A0A1A8EX70 C1BIA8 B5X8E6 H3D363 Q8K0H5 A0A3S3P410 A0A1L8DGY8 A0A1B6D4A5 A0A060YFI9 H2UQS7 A0A1A8L6I8 F7FRQ9 A0A3B1JZM9 A0A093F905 A0A1A6HQN7 G7NE05 G1SRB2 A0A093KGM3 A0A091U720 A0A087VEB8 A0A091LHN5 A0A091JJP1 A0A341CE20 A0A091V8A0 R7V2A5 A0A091TRJ8 A0A094KSU6 A0A1B0FR10 A0A151MYK0 G3U484 A0A093Q2L5 A0A2M4AM13 C1BY99 B5X692 A0A3P8XLE2 A0A091LSB3 D2HR58 V4BRI6 C1BEY2 B4F7B2 A0A091D2J5
PDB
6MZM
E-value=8.61965e-34,
Score=352
Ontologies
GO
GO:0005634
GO:0006352
GO:0003743
GO:0046982
GO:0015991
GO:0033179
GO:0016021
GO:0015078
GO:0000125
GO:0005654
GO:0051101
GO:0051260
GO:0005737
GO:0004402
GO:0006355
GO:0016578
GO:0001889
GO:0033276
GO:0003677
GO:0019899
GO:0006915
GO:0070365
GO:0016251
GO:0006367
GO:0030914
GO:0005669
GO:0070063
GO:0010468
GO:0043966
GO:0034622
GO:0000082
GO:0035264
GO:0030331
GO:0048471
GO:0017038
GO:0006605
GO:0006886
GO:0005515
GO:0003824
GO:0008152
GO:0016491
GO:0015930
PANTHER
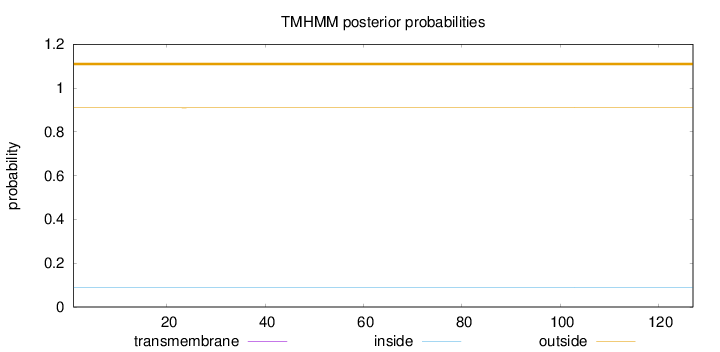
Topology
Subcellular location
Nucleus
Length:
127
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00246
Exp number, first 60 AAs:
0.00184
Total prob of N-in:
0.09035
outside
1 - 127
Population Genetic Test Statistics
Pi
178.563315
Theta
42.101337
Tajima's D
2.114181
CLR
0.147034
CSRT
0.901704914754262
Interpretation
Uncertain
