Pre Gene Modal
BGIBMGA014598
Annotation
PREDICTED:_THAP_domain-containing_protein_4-like_isoform_X1_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.963 Nuclear Reliability : 1.659
Sequence
CDS
ATGTCGAAGATTCATGAAGCTTTGGCGCCCATTTCCTGGTTGGAAGGCCGTTGGTCCACAACTGATGGCAGAGGATATTATCCGAACATACCAGACTTTACATATCATGAAGACTTGGAGTTTCTCTGCATTGGACAGCCCATGTACAATTTCCTATCGACGTCAAGGCACCCTGAAAAGCAGACGCCGATGCATCAGGAACGAGGCTTCCTTCGCATCAAGCCTGGGACAAATGAGCTGTCTTTCGTCGTGAGCCACAACTTTGGCCTGACTTCACTCGAAGAGGGACTGTGTGAGACAGAGACACATCAAGTAATTTTAGAAACAGTTAATTTATCTAGAATTTCATTTGCTAAACCACCCTTTGTGAAAAAAATTAAGAGGGTTTTCAAACTACTGAGTGATGATCACTTGGAAGCAACATTGTATATGGAAACTGATACTACACCAATGAGTGAACATTTGAAAGCAGTTTACAAAAAAATTCAATAG
Protein
MSKIHEALAPISWLEGRWSTTDGRGYYPNIPDFTYHEDLEFLCIGQPMYNFLSTSRHPEKQTPMHQERGFLRIKPGTNELSFVVSHNFGLTSLEEGLCETETHQVILETVNLSRISFAKPPFVKKIKRVFKLLSDDHLEATLYMETDTTPMSEHLKAVYKKIQ
Summary
Uniprot
H9JYH8
A0A2A4IYY6
A0A2W1BKL0
A0A2H1V841
A0A212EUM5
A0A194Q0S8
+ More
A0A194R5H7 A0A2H1WBF8 A0A182XKH0 A0A182LCU8 A0A182PK58 A0A0L7KFK4 U5ESU6 A0A182VXN8 A0A182JZN5 A0A182V5Z3 A0A182HJH4 Q7Q0H8 A0A182U0V7 A0A1B6DRQ0 A0A1Q3F3N3 A0A182SS04 B0X2P7 W5JQP7 A0A182LW67 A0A2M4AWS4 A0A2M4C1X5 A0A2M4C1Z1 A0A182NL82 A0A182FAW2 S4P5L0 A0A2T7NAS2 D6WTT0 T1E906 A0A2M3ZD65 A0A182YIF5 A0A1B6L8V4 A0A336MLJ5 A0A1B6HAA9 A0A1B6GE48 A0A182Q6J4 A0A026WZW3 K7IR91 A0A1S3I2B4 A0A0B6Z6E5 A0A1Q3F3R2 A0A1S3I2B6 A0A2D0SVY2 A0A3Q3JY83 A0A1A8L3G8 A0A1A8QE21 A0A1J1HJQ0 E1ZV30 A0A3Q3M6M1 A0A3Q2CEG8 A0A1A8NUV0 A0A1A8C4J9 A0A1A8DZ19 A0A146VBB3 A0A146VAI4 A0A1A8JXT5 A0A3B4ALT2 A0A3Q2CEP0 A0A3B4U3R5 A0A1A8V3Y1 A0A158P257 F4W6E2 W5KF72 A0A0B6Z3U0 A0A3B4XCJ1 A0A3B4ZW66 A0A3Q0S927 A0A1Y1LC58 A0A3Q1GZL8 V9LCX4 V9LCE7 A0A3Q1B370 A0A3P8SE00 A0A3B3CV70 A0A087XVX4 A0A3B3W1R2 A0A151X022 A0A1A7XIM8 A0A3B1J996 A0A1A8EVX6 A0A315VS22 A0A3Q3FRX1 A0A146NKC9 A0A0S7I7N7 A0A2I4D3G1 A0A3Q3FXQ8 A0A3Q1I8N1 A0A3Q2YBM8 A0A3B3YJA1 A0A3B3RDT2 A0A3B5KSC4 A0A0P7V5H6 A0A1U8C387 M4ANG2
A0A194R5H7 A0A2H1WBF8 A0A182XKH0 A0A182LCU8 A0A182PK58 A0A0L7KFK4 U5ESU6 A0A182VXN8 A0A182JZN5 A0A182V5Z3 A0A182HJH4 Q7Q0H8 A0A182U0V7 A0A1B6DRQ0 A0A1Q3F3N3 A0A182SS04 B0X2P7 W5JQP7 A0A182LW67 A0A2M4AWS4 A0A2M4C1X5 A0A2M4C1Z1 A0A182NL82 A0A182FAW2 S4P5L0 A0A2T7NAS2 D6WTT0 T1E906 A0A2M3ZD65 A0A182YIF5 A0A1B6L8V4 A0A336MLJ5 A0A1B6HAA9 A0A1B6GE48 A0A182Q6J4 A0A026WZW3 K7IR91 A0A1S3I2B4 A0A0B6Z6E5 A0A1Q3F3R2 A0A1S3I2B6 A0A2D0SVY2 A0A3Q3JY83 A0A1A8L3G8 A0A1A8QE21 A0A1J1HJQ0 E1ZV30 A0A3Q3M6M1 A0A3Q2CEG8 A0A1A8NUV0 A0A1A8C4J9 A0A1A8DZ19 A0A146VBB3 A0A146VAI4 A0A1A8JXT5 A0A3B4ALT2 A0A3Q2CEP0 A0A3B4U3R5 A0A1A8V3Y1 A0A158P257 F4W6E2 W5KF72 A0A0B6Z3U0 A0A3B4XCJ1 A0A3B4ZW66 A0A3Q0S927 A0A1Y1LC58 A0A3Q1GZL8 V9LCX4 V9LCE7 A0A3Q1B370 A0A3P8SE00 A0A3B3CV70 A0A087XVX4 A0A3B3W1R2 A0A151X022 A0A1A7XIM8 A0A3B1J996 A0A1A8EVX6 A0A315VS22 A0A3Q3FRX1 A0A146NKC9 A0A0S7I7N7 A0A2I4D3G1 A0A3Q3FXQ8 A0A3Q1I8N1 A0A3Q2YBM8 A0A3B3YJA1 A0A3B3RDT2 A0A3B5KSC4 A0A0P7V5H6 A0A1U8C387 M4ANG2
Pubmed
EMBL
BABH01043559
NWSH01004536
PCG64970.1
PCG64971.1
KZ150209
PZC72213.1
+ More
ODYU01001160 SOQ36981.1 AGBW02012340 OWR45189.1 KQ459580 KPI99187.1 KQ460685 KPJ12769.1 ODYU01007535 SOQ50390.1 JTDY01010031 KOB61649.1 GANO01004673 JAB55198.1 APCN01002167 AAAB01008982 EAA14625.2 GEDC01015663 GEDC01009015 JAS21635.1 JAS28283.1 GFDL01012892 JAV22153.1 DS232294 EDS39371.1 ADMH02000775 ETN65079.1 AXCM01011237 GGFK01011922 MBW45243.1 GGFJ01010144 MBW59285.1 GGFJ01010143 MBW59284.1 GAIX01008547 JAA84013.1 PZQS01000014 PVD18273.1 KQ971352 EFA06278.1 GAMD01002283 JAA99307.1 GGFM01005700 MBW26451.1 GEBQ01019834 JAT20143.1 UFQT01001605 SSX31146.1 GECU01036070 GECU01016545 JAS71636.1 JAS91161.1 GECZ01009044 JAS60725.1 AXCN02000111 KK107054 QOIP01000004 EZA61567.1 RLU23283.1 HACG01016435 CEK63300.1 GFDL01012850 JAV22195.1 HAEF01001052 SBR38434.1 HAEI01005016 SBR91399.1 CVRI01000006 CRK88253.1 GL434395 EFN74959.1 HAEH01003896 SBR72497.1 HADZ01009684 SBP73625.1 HAEA01010021 SBQ38501.1 GCES01072106 JAR14217.1 GCES01072105 JAR14218.1 HAED01019531 HAEE01004796 SBR24816.1 HADY01006633 HAEJ01013501 SBS53958.1 ADTU01006991 GL887707 EGI70277.1 HACG01016434 CEK63299.1 GEZM01060136 JAV71252.1 JW877652 AFP10169.1 JW877540 AFP10057.1 AYCK01007807 KQ982625 KYQ53480.1 HADW01016517 SBP17917.1 HAEB01004768 HAEC01009496 SBQ51295.1 NHOQ01001318 PWA25165.1 GCES01154029 JAQ32293.1 GBYX01433547 JAO47773.1 JARO02000796 KPP77375.1
ODYU01001160 SOQ36981.1 AGBW02012340 OWR45189.1 KQ459580 KPI99187.1 KQ460685 KPJ12769.1 ODYU01007535 SOQ50390.1 JTDY01010031 KOB61649.1 GANO01004673 JAB55198.1 APCN01002167 AAAB01008982 EAA14625.2 GEDC01015663 GEDC01009015 JAS21635.1 JAS28283.1 GFDL01012892 JAV22153.1 DS232294 EDS39371.1 ADMH02000775 ETN65079.1 AXCM01011237 GGFK01011922 MBW45243.1 GGFJ01010144 MBW59285.1 GGFJ01010143 MBW59284.1 GAIX01008547 JAA84013.1 PZQS01000014 PVD18273.1 KQ971352 EFA06278.1 GAMD01002283 JAA99307.1 GGFM01005700 MBW26451.1 GEBQ01019834 JAT20143.1 UFQT01001605 SSX31146.1 GECU01036070 GECU01016545 JAS71636.1 JAS91161.1 GECZ01009044 JAS60725.1 AXCN02000111 KK107054 QOIP01000004 EZA61567.1 RLU23283.1 HACG01016435 CEK63300.1 GFDL01012850 JAV22195.1 HAEF01001052 SBR38434.1 HAEI01005016 SBR91399.1 CVRI01000006 CRK88253.1 GL434395 EFN74959.1 HAEH01003896 SBR72497.1 HADZ01009684 SBP73625.1 HAEA01010021 SBQ38501.1 GCES01072106 JAR14217.1 GCES01072105 JAR14218.1 HAED01019531 HAEE01004796 SBR24816.1 HADY01006633 HAEJ01013501 SBS53958.1 ADTU01006991 GL887707 EGI70277.1 HACG01016434 CEK63299.1 GEZM01060136 JAV71252.1 JW877652 AFP10169.1 JW877540 AFP10057.1 AYCK01007807 KQ982625 KYQ53480.1 HADW01016517 SBP17917.1 HAEB01004768 HAEC01009496 SBQ51295.1 NHOQ01001318 PWA25165.1 GCES01154029 JAQ32293.1 GBYX01433547 JAO47773.1 JARO02000796 KPP77375.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000076407
+ More
UP000075882 UP000075885 UP000037510 UP000075920 UP000075881 UP000075903 UP000075840 UP000007062 UP000075902 UP000075901 UP000002320 UP000000673 UP000075883 UP000075884 UP000069272 UP000245119 UP000007266 UP000076408 UP000075886 UP000053097 UP000279307 UP000002358 UP000085678 UP000221080 UP000261600 UP000183832 UP000000311 UP000261640 UP000265020 UP000265000 UP000261520 UP000261420 UP000005205 UP000007755 UP000018467 UP000261360 UP000261400 UP000261340 UP000257200 UP000257160 UP000265080 UP000261560 UP000028760 UP000261500 UP000075809 UP000264800 UP000192220 UP000261660 UP000265040 UP000264820 UP000261480 UP000261540 UP000261380 UP000034805 UP000189706 UP000002852
UP000075882 UP000075885 UP000037510 UP000075920 UP000075881 UP000075903 UP000075840 UP000007062 UP000075902 UP000075901 UP000002320 UP000000673 UP000075883 UP000075884 UP000069272 UP000245119 UP000007266 UP000076408 UP000075886 UP000053097 UP000279307 UP000002358 UP000085678 UP000221080 UP000261600 UP000183832 UP000000311 UP000261640 UP000265020 UP000265000 UP000261520 UP000261420 UP000005205 UP000007755 UP000018467 UP000261360 UP000261400 UP000261340 UP000257200 UP000257160 UP000265080 UP000261560 UP000028760 UP000261500 UP000075809 UP000264800 UP000192220 UP000261660 UP000265040 UP000264820 UP000261480 UP000261540 UP000261380 UP000034805 UP000189706 UP000002852
PRIDE
SUPFAM
SSF50814
SSF50814
Gene 3D
ProteinModelPortal
H9JYH8
A0A2A4IYY6
A0A2W1BKL0
A0A2H1V841
A0A212EUM5
A0A194Q0S8
+ More
A0A194R5H7 A0A2H1WBF8 A0A182XKH0 A0A182LCU8 A0A182PK58 A0A0L7KFK4 U5ESU6 A0A182VXN8 A0A182JZN5 A0A182V5Z3 A0A182HJH4 Q7Q0H8 A0A182U0V7 A0A1B6DRQ0 A0A1Q3F3N3 A0A182SS04 B0X2P7 W5JQP7 A0A182LW67 A0A2M4AWS4 A0A2M4C1X5 A0A2M4C1Z1 A0A182NL82 A0A182FAW2 S4P5L0 A0A2T7NAS2 D6WTT0 T1E906 A0A2M3ZD65 A0A182YIF5 A0A1B6L8V4 A0A336MLJ5 A0A1B6HAA9 A0A1B6GE48 A0A182Q6J4 A0A026WZW3 K7IR91 A0A1S3I2B4 A0A0B6Z6E5 A0A1Q3F3R2 A0A1S3I2B6 A0A2D0SVY2 A0A3Q3JY83 A0A1A8L3G8 A0A1A8QE21 A0A1J1HJQ0 E1ZV30 A0A3Q3M6M1 A0A3Q2CEG8 A0A1A8NUV0 A0A1A8C4J9 A0A1A8DZ19 A0A146VBB3 A0A146VAI4 A0A1A8JXT5 A0A3B4ALT2 A0A3Q2CEP0 A0A3B4U3R5 A0A1A8V3Y1 A0A158P257 F4W6E2 W5KF72 A0A0B6Z3U0 A0A3B4XCJ1 A0A3B4ZW66 A0A3Q0S927 A0A1Y1LC58 A0A3Q1GZL8 V9LCX4 V9LCE7 A0A3Q1B370 A0A3P8SE00 A0A3B3CV70 A0A087XVX4 A0A3B3W1R2 A0A151X022 A0A1A7XIM8 A0A3B1J996 A0A1A8EVX6 A0A315VS22 A0A3Q3FRX1 A0A146NKC9 A0A0S7I7N7 A0A2I4D3G1 A0A3Q3FXQ8 A0A3Q1I8N1 A0A3Q2YBM8 A0A3B3YJA1 A0A3B3RDT2 A0A3B5KSC4 A0A0P7V5H6 A0A1U8C387 M4ANG2
A0A194R5H7 A0A2H1WBF8 A0A182XKH0 A0A182LCU8 A0A182PK58 A0A0L7KFK4 U5ESU6 A0A182VXN8 A0A182JZN5 A0A182V5Z3 A0A182HJH4 Q7Q0H8 A0A182U0V7 A0A1B6DRQ0 A0A1Q3F3N3 A0A182SS04 B0X2P7 W5JQP7 A0A182LW67 A0A2M4AWS4 A0A2M4C1X5 A0A2M4C1Z1 A0A182NL82 A0A182FAW2 S4P5L0 A0A2T7NAS2 D6WTT0 T1E906 A0A2M3ZD65 A0A182YIF5 A0A1B6L8V4 A0A336MLJ5 A0A1B6HAA9 A0A1B6GE48 A0A182Q6J4 A0A026WZW3 K7IR91 A0A1S3I2B4 A0A0B6Z6E5 A0A1Q3F3R2 A0A1S3I2B6 A0A2D0SVY2 A0A3Q3JY83 A0A1A8L3G8 A0A1A8QE21 A0A1J1HJQ0 E1ZV30 A0A3Q3M6M1 A0A3Q2CEG8 A0A1A8NUV0 A0A1A8C4J9 A0A1A8DZ19 A0A146VBB3 A0A146VAI4 A0A1A8JXT5 A0A3B4ALT2 A0A3Q2CEP0 A0A3B4U3R5 A0A1A8V3Y1 A0A158P257 F4W6E2 W5KF72 A0A0B6Z3U0 A0A3B4XCJ1 A0A3B4ZW66 A0A3Q0S927 A0A1Y1LC58 A0A3Q1GZL8 V9LCX4 V9LCE7 A0A3Q1B370 A0A3P8SE00 A0A3B3CV70 A0A087XVX4 A0A3B3W1R2 A0A151X022 A0A1A7XIM8 A0A3B1J996 A0A1A8EVX6 A0A315VS22 A0A3Q3FRX1 A0A146NKC9 A0A0S7I7N7 A0A2I4D3G1 A0A3Q3FXQ8 A0A3Q1I8N1 A0A3Q2YBM8 A0A3B3YJA1 A0A3B3RDT2 A0A3B5KSC4 A0A0P7V5H6 A0A1U8C387 M4ANG2
PDB
3IA8
E-value=1.06559e-26,
Score=293
Ontologies
KEGG
GO
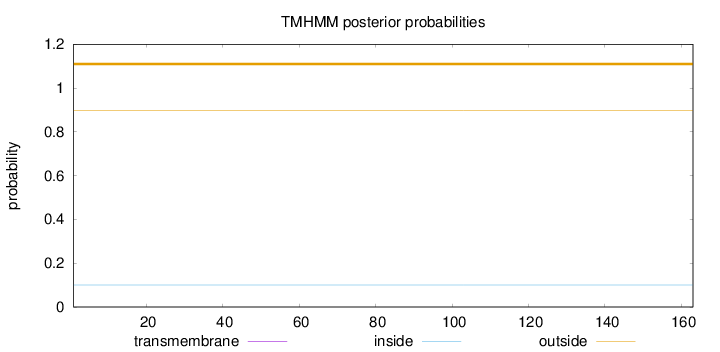
Topology
Length:
163
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00059
Exp number, first 60 AAs:
0.00059
Total prob of N-in:
0.10100
outside
1 - 163
Population Genetic Test Statistics
Pi
245.10202
Theta
220.992435
Tajima's D
0.286135
CLR
0
CSRT
0.450427478626069
Interpretation
Uncertain
