Pre Gene Modal
BGIBMGA009625
Annotation
PREDICTED:_RNA-binding_protein_25_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.353
Sequence
CDS
ATGAGGCCACATAAAGAGAAAAAAGACAAAGAAAAAGCGGACAAAGAAAAGAAGAAGAACAGTGACGGAGACAAGGAGAACAAGAGCTCCGAGGAGAAACGGAAACACATCAAGAGTCTCATCGACAAGATCCCCACGCAGAAGGAACAACTGTTCCAGTACAAACTGGACGCCTCGCAGATCGATAACACGCTCATGGAGAAGAAGATACGTCCATGGATCAACAAGAAGATCATAGAGTACATCGGCGAGCCGGAGCCCACTCTCGTCGACTTCATCTGCAGCAAAGTGCTCGCCGGCTCCGCGCCCCAGGGAGTGCTCGACGACGTGCAGATGGTGCTGGACGAGGAGGCGGAGGTGTTCGTGGTGAAGATGTGGCGGCTGCTCATCTACGAGCTGGAGGCCAAGCGCGCCGGCCTGCACAAGTGA
Protein
MRPHKEKKDKEKADKEKKKNSDGDKENKSSEEKRKHIKSLIDKIPTQKEQLFQYKLDASQIDNTLMEKKIRPWINKKIIEYIGEPEPTLVDFICSKVLAGSAPQGVLDDVQMVLDEEAEVFVVKMWRLLIYELEAKRAGLHK
Summary
Uniprot
H9JJC4
A0A2H1WXP0
A0A3S2TAK1
A0A2W1B0J3
A0A1E1W423
A0A0N0PCP6
+ More
A0A212ENX9 A0A0K8W9T8 A0A1B0CAP9 A0A1B6IFW1 A0A0A9XI46 A0A2J7R980 A0A1L8DHA8 A0A151WMA5 A0A0J7L828 A0A195CAC2 E9J569 A0A026WPD8 A0A2J7R984 A0A154PDG6 E2C9R8 A0A0L7RHS2 T1P8W8 A0A195B0T1 A0A0C9QZH4 A0A088AGZ0 A0A2A3ESM0 A0A195EU96 A0A158NR25 A0A195E6L0 F4WPA0 A0A310SAB0 A0A1B6L094 A0A2P8XVW8 A0A1B6FZ11 A0A023FGJ6 A0A2J7R986 A0A0K8U173 A0A1A9USU6 A0A1I8P2M5 A0A1A9ZQG6 A0A1B0FFE4 A0A1B0BCU4 A0A1I8MAX5 A0A1A9YJ90 A0A1A9W900 A0A0L0C622 W8C943 A0A1Y1L015 A0A034W599 A0A0C9PX77 B4H4G4 B4R506 A0A232FKT1 B4I0M0 K7IQ34 B5DLW5 A0A3B0K674 A0A0P4VP07 A0A0A9XF17 Q9V3Y5 A0A0A9WNG6 E2ATS2 A0A1B6IXZ0 A0A3B0K8G8 A0A069DZK1 B4M805 A0A2J7R987 V5FXN4 B3MRX5 A0A1W4VTH4 A0A1B6C558 B4L643 B4JKI3 A0A1B6EE52 A0A336MEU6 A0A336MGC6 A0A0M3QZS5 B4MSG4 E0VC25 N6SXF3 B3NV67 A0A1Y1L023 A0A1Y1L4U4 A0A3Q0IXJ1 A0A084WTV1 A0A0R1E968 B4PZC9 U4URA2 A0A023EY32 A0A182RE00 U5EZV2 A0A182TWZ4 A0A182JHC4 A0A182G6U8 A0A182N9V5 A0A182KZF9 A0A182W3K3 A0A182Q7B4 B0WNI2
A0A212ENX9 A0A0K8W9T8 A0A1B0CAP9 A0A1B6IFW1 A0A0A9XI46 A0A2J7R980 A0A1L8DHA8 A0A151WMA5 A0A0J7L828 A0A195CAC2 E9J569 A0A026WPD8 A0A2J7R984 A0A154PDG6 E2C9R8 A0A0L7RHS2 T1P8W8 A0A195B0T1 A0A0C9QZH4 A0A088AGZ0 A0A2A3ESM0 A0A195EU96 A0A158NR25 A0A195E6L0 F4WPA0 A0A310SAB0 A0A1B6L094 A0A2P8XVW8 A0A1B6FZ11 A0A023FGJ6 A0A2J7R986 A0A0K8U173 A0A1A9USU6 A0A1I8P2M5 A0A1A9ZQG6 A0A1B0FFE4 A0A1B0BCU4 A0A1I8MAX5 A0A1A9YJ90 A0A1A9W900 A0A0L0C622 W8C943 A0A1Y1L015 A0A034W599 A0A0C9PX77 B4H4G4 B4R506 A0A232FKT1 B4I0M0 K7IQ34 B5DLW5 A0A3B0K674 A0A0P4VP07 A0A0A9XF17 Q9V3Y5 A0A0A9WNG6 E2ATS2 A0A1B6IXZ0 A0A3B0K8G8 A0A069DZK1 B4M805 A0A2J7R987 V5FXN4 B3MRX5 A0A1W4VTH4 A0A1B6C558 B4L643 B4JKI3 A0A1B6EE52 A0A336MEU6 A0A336MGC6 A0A0M3QZS5 B4MSG4 E0VC25 N6SXF3 B3NV67 A0A1Y1L023 A0A1Y1L4U4 A0A3Q0IXJ1 A0A084WTV1 A0A0R1E968 B4PZC9 U4URA2 A0A023EY32 A0A182RE00 U5EZV2 A0A182TWZ4 A0A182JHC4 A0A182G6U8 A0A182N9V5 A0A182KZF9 A0A182W3K3 A0A182Q7B4 B0WNI2
Pubmed
19121390
28756777
26354079
22118469
25401762
21282665
+ More
24508170 30249741 20798317 21347285 21719571 29403074 25315136 26108605 24495485 28004739 25348373 17994087 28648823 20075255 15632085 27129103 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26334808 18057021 20566863 23537049 24438588 17550304 24945155 26483478 20966253
24508170 30249741 20798317 21347285 21719571 29403074 25315136 26108605 24495485 28004739 25348373 17994087 28648823 20075255 15632085 27129103 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26334808 18057021 20566863 23537049 24438588 17550304 24945155 26483478 20966253
EMBL
BABH01036448
BABH01036449
BABH01036450
BABH01036451
BABH01036452
BABH01036453
+ More
ODYU01011816 SOQ57777.1 RSAL01003036 RVE40358.1 KZ150505 PZC70652.1 GDQN01009352 JAT81702.1 KQ460501 KPJ14181.1 AGBW02013604 OWR43184.1 GDHF01004520 JAI47794.1 AJWK01004153 GECU01021898 JAS85808.1 GBHO01034641 GBHO01034638 GBHO01034637 GBHO01034636 GBHO01024284 GBHO01024283 JAG08963.1 JAG08966.1 JAG08967.1 JAG08968.1 JAG19320.1 JAG19321.1 NEVH01006600 PNF37396.1 GFDF01008245 JAV05839.1 KQ982944 KYQ49022.1 LBMM01000293 KMR01498.1 KQ978081 KYM97136.1 GL768124 EFZ12014.1 KK107144 QOIP01000003 EZA57521.1 RLU25286.1 PNF37399.1 KQ434879 KZC09946.1 GL453897 EFN75308.1 KQ414591 KOC70296.1 KA644585 AFP59214.1 KQ976691 KYM77902.1 GBYB01006112 JAG75879.1 KZ288189 PBC34697.1 KQ981965 KYN31825.1 ADTU01023754 ADTU01023755 KQ979608 KYN20484.1 GL888243 EGI64081.1 KQ797310 OAD46973.1 GEBQ01022869 JAT17108.1 PYGN01001265 PSN36150.1 GECZ01014332 JAS55437.1 GBBK01004358 JAC20124.1 PNF37397.1 GDHF01032224 JAI20090.1 CCAG010018023 JXJN01012125 JRES01000853 KNC27730.1 GAMC01006971 JAB99584.1 GEZM01068343 GEZM01068342 GEZM01068339 GEZM01068338 GEZM01068337 GEZM01068336 GEZM01068335 GEZM01068334 GEZM01068333 GEZM01068331 JAV66993.1 GAKP01009470 JAC49482.1 GBYB01006113 JAG75880.1 CH479209 EDW32715.1 CM000366 EDX17155.1 NNAY01000067 OXU31344.1 CH480819 EDW53051.1 CH379064 EDY72512.1 OUUW01000025 SPP89634.1 GDKW01000066 JAI56529.1 GBHO01024282 GBHO01009612 GBRD01010208 JAG19322.1 JAG33992.1 JAG55616.1 AF181653 AE014298 AAD55438.1 AAF46056.1 GBHO01034640 GBHO01034633 JAG08964.1 JAG08971.1 GL442691 EFN63155.1 GECU01015996 JAS91710.1 SPP89633.1 GBGD01000500 JAC88389.1 CH940653 EDW62281.1 KRF80535.1 KRF80536.1 PNF37398.1 GALX01006051 JAB62415.1 CH902622 EDV34530.1 GEDC01028933 JAS08365.1 CH933812 EDW05839.1 CH916370 EDW00086.1 GEDC01001138 JAS36160.1 UFQT01001009 SSX28460.1 SSX28461.1 CP012528 ALC49889.1 CH963851 EDW75053.2 DS235045 EEB10931.1 APGK01052763 KB741216 ENN72454.1 CH954180 EDV45915.1 GEZM01068332 JAV67002.1 GEZM01068341 JAV66985.1 ATLV01026919 KE525420 KFB53645.1 CM000162 KRK05952.1 KRK05953.1 EDX01096.1 KRK05954.1 KB632404 ERL95038.1 GAPW01000159 JAC13439.1 GANO01000085 JAB59786.1 JXUM01148920 JXUM01148921 JXUM01148922 JXUM01148923 KQ570665 KXJ68320.1 AXCN02001201 DS232010 EDS31636.1
ODYU01011816 SOQ57777.1 RSAL01003036 RVE40358.1 KZ150505 PZC70652.1 GDQN01009352 JAT81702.1 KQ460501 KPJ14181.1 AGBW02013604 OWR43184.1 GDHF01004520 JAI47794.1 AJWK01004153 GECU01021898 JAS85808.1 GBHO01034641 GBHO01034638 GBHO01034637 GBHO01034636 GBHO01024284 GBHO01024283 JAG08963.1 JAG08966.1 JAG08967.1 JAG08968.1 JAG19320.1 JAG19321.1 NEVH01006600 PNF37396.1 GFDF01008245 JAV05839.1 KQ982944 KYQ49022.1 LBMM01000293 KMR01498.1 KQ978081 KYM97136.1 GL768124 EFZ12014.1 KK107144 QOIP01000003 EZA57521.1 RLU25286.1 PNF37399.1 KQ434879 KZC09946.1 GL453897 EFN75308.1 KQ414591 KOC70296.1 KA644585 AFP59214.1 KQ976691 KYM77902.1 GBYB01006112 JAG75879.1 KZ288189 PBC34697.1 KQ981965 KYN31825.1 ADTU01023754 ADTU01023755 KQ979608 KYN20484.1 GL888243 EGI64081.1 KQ797310 OAD46973.1 GEBQ01022869 JAT17108.1 PYGN01001265 PSN36150.1 GECZ01014332 JAS55437.1 GBBK01004358 JAC20124.1 PNF37397.1 GDHF01032224 JAI20090.1 CCAG010018023 JXJN01012125 JRES01000853 KNC27730.1 GAMC01006971 JAB99584.1 GEZM01068343 GEZM01068342 GEZM01068339 GEZM01068338 GEZM01068337 GEZM01068336 GEZM01068335 GEZM01068334 GEZM01068333 GEZM01068331 JAV66993.1 GAKP01009470 JAC49482.1 GBYB01006113 JAG75880.1 CH479209 EDW32715.1 CM000366 EDX17155.1 NNAY01000067 OXU31344.1 CH480819 EDW53051.1 CH379064 EDY72512.1 OUUW01000025 SPP89634.1 GDKW01000066 JAI56529.1 GBHO01024282 GBHO01009612 GBRD01010208 JAG19322.1 JAG33992.1 JAG55616.1 AF181653 AE014298 AAD55438.1 AAF46056.1 GBHO01034640 GBHO01034633 JAG08964.1 JAG08971.1 GL442691 EFN63155.1 GECU01015996 JAS91710.1 SPP89633.1 GBGD01000500 JAC88389.1 CH940653 EDW62281.1 KRF80535.1 KRF80536.1 PNF37398.1 GALX01006051 JAB62415.1 CH902622 EDV34530.1 GEDC01028933 JAS08365.1 CH933812 EDW05839.1 CH916370 EDW00086.1 GEDC01001138 JAS36160.1 UFQT01001009 SSX28460.1 SSX28461.1 CP012528 ALC49889.1 CH963851 EDW75053.2 DS235045 EEB10931.1 APGK01052763 KB741216 ENN72454.1 CH954180 EDV45915.1 GEZM01068332 JAV67002.1 GEZM01068341 JAV66985.1 ATLV01026919 KE525420 KFB53645.1 CM000162 KRK05952.1 KRK05953.1 EDX01096.1 KRK05954.1 KB632404 ERL95038.1 GAPW01000159 JAC13439.1 GANO01000085 JAB59786.1 JXUM01148920 JXUM01148921 JXUM01148922 JXUM01148923 KQ570665 KXJ68320.1 AXCN02001201 DS232010 EDS31636.1
Proteomes
UP000005204
UP000283053
UP000053240
UP000007151
UP000092461
UP000235965
+ More
UP000075809 UP000036403 UP000078542 UP000053097 UP000279307 UP000076502 UP000008237 UP000053825 UP000078540 UP000005203 UP000242457 UP000078541 UP000005205 UP000078492 UP000007755 UP000245037 UP000078200 UP000095300 UP000092445 UP000092444 UP000092460 UP000095301 UP000092443 UP000091820 UP000037069 UP000008744 UP000000304 UP000215335 UP000001292 UP000002358 UP000001819 UP000268350 UP000000803 UP000000311 UP000008792 UP000007801 UP000192221 UP000009192 UP000001070 UP000092553 UP000007798 UP000009046 UP000019118 UP000008711 UP000079169 UP000030765 UP000002282 UP000030742 UP000075900 UP000075902 UP000075880 UP000069940 UP000249989 UP000075884 UP000075882 UP000075920 UP000075886 UP000002320
UP000075809 UP000036403 UP000078542 UP000053097 UP000279307 UP000076502 UP000008237 UP000053825 UP000078540 UP000005203 UP000242457 UP000078541 UP000005205 UP000078492 UP000007755 UP000245037 UP000078200 UP000095300 UP000092445 UP000092444 UP000092460 UP000095301 UP000092443 UP000091820 UP000037069 UP000008744 UP000000304 UP000215335 UP000001292 UP000002358 UP000001819 UP000268350 UP000000803 UP000000311 UP000008792 UP000007801 UP000192221 UP000009192 UP000001070 UP000092553 UP000007798 UP000009046 UP000019118 UP000008711 UP000079169 UP000030765 UP000002282 UP000030742 UP000075900 UP000075902 UP000075880 UP000069940 UP000249989 UP000075884 UP000075882 UP000075920 UP000075886 UP000002320
Interpro
Gene 3D
ProteinModelPortal
H9JJC4
A0A2H1WXP0
A0A3S2TAK1
A0A2W1B0J3
A0A1E1W423
A0A0N0PCP6
+ More
A0A212ENX9 A0A0K8W9T8 A0A1B0CAP9 A0A1B6IFW1 A0A0A9XI46 A0A2J7R980 A0A1L8DHA8 A0A151WMA5 A0A0J7L828 A0A195CAC2 E9J569 A0A026WPD8 A0A2J7R984 A0A154PDG6 E2C9R8 A0A0L7RHS2 T1P8W8 A0A195B0T1 A0A0C9QZH4 A0A088AGZ0 A0A2A3ESM0 A0A195EU96 A0A158NR25 A0A195E6L0 F4WPA0 A0A310SAB0 A0A1B6L094 A0A2P8XVW8 A0A1B6FZ11 A0A023FGJ6 A0A2J7R986 A0A0K8U173 A0A1A9USU6 A0A1I8P2M5 A0A1A9ZQG6 A0A1B0FFE4 A0A1B0BCU4 A0A1I8MAX5 A0A1A9YJ90 A0A1A9W900 A0A0L0C622 W8C943 A0A1Y1L015 A0A034W599 A0A0C9PX77 B4H4G4 B4R506 A0A232FKT1 B4I0M0 K7IQ34 B5DLW5 A0A3B0K674 A0A0P4VP07 A0A0A9XF17 Q9V3Y5 A0A0A9WNG6 E2ATS2 A0A1B6IXZ0 A0A3B0K8G8 A0A069DZK1 B4M805 A0A2J7R987 V5FXN4 B3MRX5 A0A1W4VTH4 A0A1B6C558 B4L643 B4JKI3 A0A1B6EE52 A0A336MEU6 A0A336MGC6 A0A0M3QZS5 B4MSG4 E0VC25 N6SXF3 B3NV67 A0A1Y1L023 A0A1Y1L4U4 A0A3Q0IXJ1 A0A084WTV1 A0A0R1E968 B4PZC9 U4URA2 A0A023EY32 A0A182RE00 U5EZV2 A0A182TWZ4 A0A182JHC4 A0A182G6U8 A0A182N9V5 A0A182KZF9 A0A182W3K3 A0A182Q7B4 B0WNI2
A0A212ENX9 A0A0K8W9T8 A0A1B0CAP9 A0A1B6IFW1 A0A0A9XI46 A0A2J7R980 A0A1L8DHA8 A0A151WMA5 A0A0J7L828 A0A195CAC2 E9J569 A0A026WPD8 A0A2J7R984 A0A154PDG6 E2C9R8 A0A0L7RHS2 T1P8W8 A0A195B0T1 A0A0C9QZH4 A0A088AGZ0 A0A2A3ESM0 A0A195EU96 A0A158NR25 A0A195E6L0 F4WPA0 A0A310SAB0 A0A1B6L094 A0A2P8XVW8 A0A1B6FZ11 A0A023FGJ6 A0A2J7R986 A0A0K8U173 A0A1A9USU6 A0A1I8P2M5 A0A1A9ZQG6 A0A1B0FFE4 A0A1B0BCU4 A0A1I8MAX5 A0A1A9YJ90 A0A1A9W900 A0A0L0C622 W8C943 A0A1Y1L015 A0A034W599 A0A0C9PX77 B4H4G4 B4R506 A0A232FKT1 B4I0M0 K7IQ34 B5DLW5 A0A3B0K674 A0A0P4VP07 A0A0A9XF17 Q9V3Y5 A0A0A9WNG6 E2ATS2 A0A1B6IXZ0 A0A3B0K8G8 A0A069DZK1 B4M805 A0A2J7R987 V5FXN4 B3MRX5 A0A1W4VTH4 A0A1B6C558 B4L643 B4JKI3 A0A1B6EE52 A0A336MEU6 A0A336MGC6 A0A0M3QZS5 B4MSG4 E0VC25 N6SXF3 B3NV67 A0A1Y1L023 A0A1Y1L4U4 A0A3Q0IXJ1 A0A084WTV1 A0A0R1E968 B4PZC9 U4URA2 A0A023EY32 A0A182RE00 U5EZV2 A0A182TWZ4 A0A182JHC4 A0A182G6U8 A0A182N9V5 A0A182KZF9 A0A182W3K3 A0A182Q7B4 B0WNI2
PDB
3V53
E-value=1.73037e-39,
Score=402
Ontologies
GO
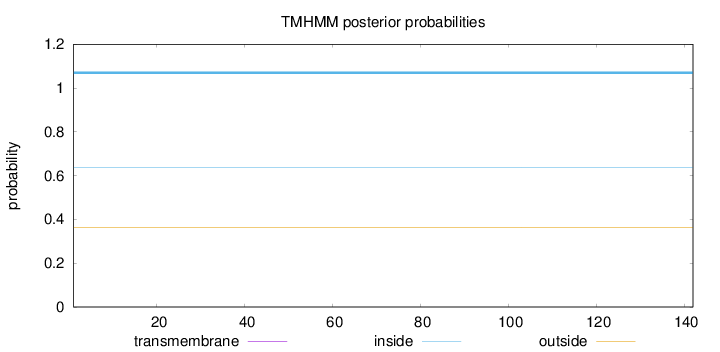
Topology
Length:
142
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000780000000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.63839
inside
1 - 142
Population Genetic Test Statistics
Pi
173.64797
Theta
72.477996
Tajima's D
1.777356
CLR
0.000881
CSRT
0.842157892105395
Interpretation
Uncertain
