Pre Gene Modal
BGIBMGA009570
Annotation
PREDICTED:_protein_IWS1_homolog_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.43
Sequence
CDS
ATGCGAGAGCCCAAGCGAGCCCGAGACGAGGGAGACGAGGCGCCCGAGGTGGAAGGGGAGGGGCGCGTGCTGGACGCGGAAGCAGCGCAGCCCGACAGCTCCGACGATGACATCGCACCCGACTCTAAGAGTGCGGAGGCGGCGGGCGGGCTGTCTGACTTCGAGGTGATGTTGGCGCGCAAGCGGGACGAGCGGCGAGGCCGGCGGCGGCGCAGGGACATCGACATCATCAACGACAACGACGACCTCATCGCGCAGCTGCTGCAGTCCATGAGGCAGGCGGCCGACGAGGACAGGGAGCTCAATCGCAAGAACCAACCCGCCGTGAAGAAGGTGTCCATGCTGAAGACGGCCATGTCGCAGCTGATCAAGAAGGACCTGCAGCTGGCCTTCCTGGAGCACAACGCGCTCAACGTGCTGTGCGACTGGCTCGCGCCCATGCCCAACCGCGCGCTGCCCTGCCTGCTCATCCGCGAGACCGTGCTCAAGCTGCTCACCGACTTTCCGCCCATAGACAAGTCGTTGCTGAAGCAGTCCGGCATCGGCAAGGCCGTGATGTACCTCTACAAGCACCCCAAGGAGACCAAGGCCAACAGAGAGCGAGCCGGCCGACTCATCTCCGAGTGGGCCAGGCCCATCTTCAACCTCTCCACAGATTTTAAAGCCATGACGCGGGAAGAGCGACAGGCACGAGATGAAGCCATGGCGGGCGGGCGGCGGCGGGATGACCCCGGACCCTCCGCCAGCGCGCAAGACGAACCCAGGGCGCGGCGGCCGGGCGAGGCGGGCTGGGTGGCGCGCGCGCGGGTGCCGCAGCCCTCCACCAAGGACTACGTGTACCGACCCAAGTCCACCTGCGACCTGGACATGTCCCGGACCACTAAGAAGAAGATGACCCGCTACGAGAAGCAAATGAAGAAGTTCGTGGATCAGAAGCGACTGAAGAGCGGCTCGCGGCGCGCGGTCGAGATCTCCATCGAGGGCCGCAAGATGGCGCTGTGA
Protein
MREPKRARDEGDEAPEVEGEGRVLDAEAAQPDSSDDDIAPDSKSAEAAGGLSDFEVMLARKRDERRGRRRRRDIDIINDNDDLIAQLLQSMRQAADEDRELNRKNQPAVKKVSMLKTAMSQLIKKDLQLAFLEHNALNVLCDWLAPMPNRALPCLLIRETVLKLLTDFPPIDKSLLKQSGIGKAVMYLYKHPKETKANRERAGRLISEWARPIFNLSTDFKAMTREERQARDEAMAGGRRRDDPGPSASAQDEPRARRPGEAGWVARARVPQPSTKDYVYRPKSTCDLDMSRTTKKKMTRYEKQMKKFVDQKRLKSGSRRAVEISIEGRKMAL
Summary
Uniprot
H9JJ69
A0A2H1WXX5
A0A2W1B0I9
A0A212ENY7
S4PTI3
U5EK72
+ More
A0A336MHC0 A0A336M8M2 A0A2M4ACU7 A0A2M4ACN3 A0A2M4BF49 W5JVZ9 A0A182FJW3 A0A2M3ZGY9 A0A2M3ZGV1 B0WWT3 A0A1B0GJC1 A0A182YCS7 A0A2M4BDS4 A0A2M4BDN3 A0A182STD2 A0A1L8DW32 A0A1L8DW35 A0A1S4JVJ2 A0A084VX58 A0A1Q3FW62 A0A023EUD7 A0A182RKY1 A0A1J1IV32 A0A182MF06 A0A182IXL1 A0A1I8MFH2 A0A182H9C6 Q7QFC0 A0A1Y9J009 A0A182K826 Q4V6M6 A0A1I8Q5V5 B4F5R2 A0A0K8TYT0 A0A182QR47 B4F5R3 A0A034WE83 A0A0A1WM66 Q29GE4 B4HB72 B4F5S4 B4IF53 A0A182PT47 A0A182W5T1 A0A0P6J688 A0A3B0KE70 W8C6N5 A0A1A9YGV9 Q179S2 A0A1B0FM02 A0A1A9X010 A0A1B0AJ06 B9EQY8 A0A1B0AT06 A0A1A9UV21 A0A0L0BUP2 A0A1W4WDI3 B4JJD5 B3N0T6 A8JV07 A0A0Q5TK49 A0A182N0K8 A0A0A9XTF3 B4L868 E2AX83 A0A1B6CVX6 B4NCM6 E0VVM6 A0A1B6IAK4 A0A1B6G047 A0A3L8DFW1 D6WHW2 E2BN92 A0A158NVV9 A0A1Y1LHS4 A0A154PII2 A0A1B6L9I5 A0A195ET90 A0A084WR77 A0A0M4EHN9 A0A2R7WAG7 A0A0P4VPV5 A0A087ZTS6 A0A2M4ADQ2 A0A0M8ZTJ4 A0A026WH59 A0A2A3EDY3 A0A3Q0JL82 A0A067QQ33 A0A0J7KCP4 A0A2J7QM59 A0A023F1G4 A0A0V0G744 A0A023F3K2
A0A336MHC0 A0A336M8M2 A0A2M4ACU7 A0A2M4ACN3 A0A2M4BF49 W5JVZ9 A0A182FJW3 A0A2M3ZGY9 A0A2M3ZGV1 B0WWT3 A0A1B0GJC1 A0A182YCS7 A0A2M4BDS4 A0A2M4BDN3 A0A182STD2 A0A1L8DW32 A0A1L8DW35 A0A1S4JVJ2 A0A084VX58 A0A1Q3FW62 A0A023EUD7 A0A182RKY1 A0A1J1IV32 A0A182MF06 A0A182IXL1 A0A1I8MFH2 A0A182H9C6 Q7QFC0 A0A1Y9J009 A0A182K826 Q4V6M6 A0A1I8Q5V5 B4F5R2 A0A0K8TYT0 A0A182QR47 B4F5R3 A0A034WE83 A0A0A1WM66 Q29GE4 B4HB72 B4F5S4 B4IF53 A0A182PT47 A0A182W5T1 A0A0P6J688 A0A3B0KE70 W8C6N5 A0A1A9YGV9 Q179S2 A0A1B0FM02 A0A1A9X010 A0A1B0AJ06 B9EQY8 A0A1B0AT06 A0A1A9UV21 A0A0L0BUP2 A0A1W4WDI3 B4JJD5 B3N0T6 A8JV07 A0A0Q5TK49 A0A182N0K8 A0A0A9XTF3 B4L868 E2AX83 A0A1B6CVX6 B4NCM6 E0VVM6 A0A1B6IAK4 A0A1B6G047 A0A3L8DFW1 D6WHW2 E2BN92 A0A158NVV9 A0A1Y1LHS4 A0A154PII2 A0A1B6L9I5 A0A195ET90 A0A084WR77 A0A0M4EHN9 A0A2R7WAG7 A0A0P4VPV5 A0A087ZTS6 A0A2M4ADQ2 A0A0M8ZTJ4 A0A026WH59 A0A2A3EDY3 A0A3Q0JL82 A0A067QQ33 A0A0J7KCP4 A0A2J7QM59 A0A023F1G4 A0A0V0G744 A0A023F3K2
Pubmed
19121390
28756777
22118469
23622113
20920257
23761445
+ More
25244985 24438588 24945155 25315136 26483478 12364791 14747013 17210077 18477586 19126864 25348373 25830018 15632085 17994087 26999592 24495485 17510324 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25401762 20798317 20566863 30249741 18362917 19820115 21347285 28004739 27129103 24508170 24845553 25474469
25244985 24438588 24945155 25315136 26483478 12364791 14747013 17210077 18477586 19126864 25348373 25830018 15632085 17994087 26999592 24495485 17510324 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25401762 20798317 20566863 30249741 18362917 19820115 21347285 28004739 27129103 24508170 24845553 25474469
EMBL
BABH01036445
ODYU01011913
SOQ57925.1
KZ150505
PZC70649.1
AGBW02013604
+ More
OWR43186.1 GAIX01011988 JAA80572.1 GANO01001920 JAB57951.1 UFQT01001272 SSX29792.1 UFQT01000555 SSX25249.1 GGFK01005294 MBW38615.1 GGFK01005220 MBW38541.1 GGFJ01002482 MBW51623.1 ADMH02000129 ETN67663.1 GGFM01007043 MBW27794.1 GGFM01007046 MBW27797.1 DS232152 EDS36194.1 AJWK01020126 GGFJ01002012 MBW51153.1 GGFJ01002011 MBW51152.1 GFDF01003434 JAV10650.1 GFDF01003433 JAV10651.1 ATLV01017849 KE525195 KFB42552.1 GFDL01003195 JAV31850.1 GAPW01000573 JAC13025.1 CVRI01000061 CRL04115.1 AXCM01000769 JXUM01029965 KQ560869 KXJ80554.1 AAAB01008846 EAA06643.5 BT022280 AAY54696.1 AM999148 CAQ53510.1 GDHF01032878 JAI19436.1 AXCN02002228 AM999149 AM999150 AM999151 AM999152 AM999153 AM999154 AM999155 AM999156 AM999157 AM999158 AM999159 FM246264 FM246265 FM246266 FM246267 FM246268 FM246269 FM246270 FM246271 FM246272 CAQ53511.1 CAQ53512.1 CAQ53513.1 CAQ53514.1 CAQ53515.1 CAQ53516.1 CAQ53517.1 CAQ53518.1 CAQ53519.1 CAQ53520.1 CAQ53521.1 CAR94190.1 CAR94191.1 CAR94192.1 CAR94193.1 CAR94194.1 CAR94195.1 CAR94196.1 CAR94197.1 CAR94198.1 GAKP01005953 JAC52999.1 GBXI01014677 JAC99614.1 CH379064 EAL32165.2 CH479248 EDW37884.1 AM999160 CAQ53522.1 CH480832 EDW46107.1 GDUN01000198 JAN95721.1 OUUW01000011 SPP86600.1 GAMC01004139 JAC02417.1 CH477346 EAT42999.1 CCAG010015359 BT058063 ACM78505.1 JXJN01003090 JRES01001303 KNC23751.1 CH916370 EDV99687.1 CH902644 EDV34815.2 AE014298 ABW09431.1 AGB95446.1 CH954180 KQS30230.1 GBHO01020455 GBRD01017753 JAG23149.1 JAG48074.1 CH933814 EDW05643.2 GL443520 EFN61966.1 GEDC01019813 JAS17485.1 CH964239 EDW82585.2 DS235812 EEB17432.1 GECU01023757 JAS83949.1 GECZ01013955 JAS55814.1 QOIP01000009 RLU18799.1 KQ971321 EFA00703.2 GL449385 EFN82854.1 ADTU01027542 GEZM01061276 JAV70577.1 KQ434899 KZC11000.1 GEBQ01019608 JAT20369.1 KQ981993 KYN31099.1 ATLV01025961 KE525403 KFB52721.1 CP012528 ALC48191.1 KK854459 PTY16020.1 GDKW01003388 JAI53207.1 GGFK01005590 MBW38911.1 KQ435851 KOX70845.1 KK107231 EZA55006.1 KZ288291 PBC29221.1 KK853149 KDR10670.1 LBMM01009353 KMQ88188.1 NEVH01013216 PNF29680.1 GBBI01003397 JAC15315.1 GECL01002335 JAP03789.1 GBBI01003273 JAC15439.1
OWR43186.1 GAIX01011988 JAA80572.1 GANO01001920 JAB57951.1 UFQT01001272 SSX29792.1 UFQT01000555 SSX25249.1 GGFK01005294 MBW38615.1 GGFK01005220 MBW38541.1 GGFJ01002482 MBW51623.1 ADMH02000129 ETN67663.1 GGFM01007043 MBW27794.1 GGFM01007046 MBW27797.1 DS232152 EDS36194.1 AJWK01020126 GGFJ01002012 MBW51153.1 GGFJ01002011 MBW51152.1 GFDF01003434 JAV10650.1 GFDF01003433 JAV10651.1 ATLV01017849 KE525195 KFB42552.1 GFDL01003195 JAV31850.1 GAPW01000573 JAC13025.1 CVRI01000061 CRL04115.1 AXCM01000769 JXUM01029965 KQ560869 KXJ80554.1 AAAB01008846 EAA06643.5 BT022280 AAY54696.1 AM999148 CAQ53510.1 GDHF01032878 JAI19436.1 AXCN02002228 AM999149 AM999150 AM999151 AM999152 AM999153 AM999154 AM999155 AM999156 AM999157 AM999158 AM999159 FM246264 FM246265 FM246266 FM246267 FM246268 FM246269 FM246270 FM246271 FM246272 CAQ53511.1 CAQ53512.1 CAQ53513.1 CAQ53514.1 CAQ53515.1 CAQ53516.1 CAQ53517.1 CAQ53518.1 CAQ53519.1 CAQ53520.1 CAQ53521.1 CAR94190.1 CAR94191.1 CAR94192.1 CAR94193.1 CAR94194.1 CAR94195.1 CAR94196.1 CAR94197.1 CAR94198.1 GAKP01005953 JAC52999.1 GBXI01014677 JAC99614.1 CH379064 EAL32165.2 CH479248 EDW37884.1 AM999160 CAQ53522.1 CH480832 EDW46107.1 GDUN01000198 JAN95721.1 OUUW01000011 SPP86600.1 GAMC01004139 JAC02417.1 CH477346 EAT42999.1 CCAG010015359 BT058063 ACM78505.1 JXJN01003090 JRES01001303 KNC23751.1 CH916370 EDV99687.1 CH902644 EDV34815.2 AE014298 ABW09431.1 AGB95446.1 CH954180 KQS30230.1 GBHO01020455 GBRD01017753 JAG23149.1 JAG48074.1 CH933814 EDW05643.2 GL443520 EFN61966.1 GEDC01019813 JAS17485.1 CH964239 EDW82585.2 DS235812 EEB17432.1 GECU01023757 JAS83949.1 GECZ01013955 JAS55814.1 QOIP01000009 RLU18799.1 KQ971321 EFA00703.2 GL449385 EFN82854.1 ADTU01027542 GEZM01061276 JAV70577.1 KQ434899 KZC11000.1 GEBQ01019608 JAT20369.1 KQ981993 KYN31099.1 ATLV01025961 KE525403 KFB52721.1 CP012528 ALC48191.1 KK854459 PTY16020.1 GDKW01003388 JAI53207.1 GGFK01005590 MBW38911.1 KQ435851 KOX70845.1 KK107231 EZA55006.1 KZ288291 PBC29221.1 KK853149 KDR10670.1 LBMM01009353 KMQ88188.1 NEVH01013216 PNF29680.1 GBBI01003397 JAC15315.1 GECL01002335 JAP03789.1 GBBI01003273 JAC15439.1
Proteomes
UP000005204
UP000007151
UP000000673
UP000069272
UP000002320
UP000092461
+ More
UP000076408 UP000075901 UP000030765 UP000075900 UP000183832 UP000075883 UP000075880 UP000095301 UP000069940 UP000249989 UP000007062 UP000076407 UP000075881 UP000095300 UP000075886 UP000001819 UP000008744 UP000001292 UP000075885 UP000075920 UP000268350 UP000092443 UP000008820 UP000092444 UP000091820 UP000092445 UP000092460 UP000078200 UP000037069 UP000192221 UP000001070 UP000007801 UP000000803 UP000008711 UP000075884 UP000009192 UP000000311 UP000007798 UP000009046 UP000279307 UP000007266 UP000008237 UP000005205 UP000076502 UP000078541 UP000092553 UP000005203 UP000053105 UP000053097 UP000242457 UP000079169 UP000027135 UP000036403 UP000235965
UP000076408 UP000075901 UP000030765 UP000075900 UP000183832 UP000075883 UP000075880 UP000095301 UP000069940 UP000249989 UP000007062 UP000076407 UP000075881 UP000095300 UP000075886 UP000001819 UP000008744 UP000001292 UP000075885 UP000075920 UP000268350 UP000092443 UP000008820 UP000092444 UP000091820 UP000092445 UP000092460 UP000078200 UP000037069 UP000192221 UP000001070 UP000007801 UP000000803 UP000008711 UP000075884 UP000009192 UP000000311 UP000007798 UP000009046 UP000279307 UP000007266 UP000008237 UP000005205 UP000076502 UP000078541 UP000092553 UP000005203 UP000053105 UP000053097 UP000242457 UP000079169 UP000027135 UP000036403 UP000235965
SUPFAM
SSF47676
SSF47676
Gene 3D
ProteinModelPortal
H9JJ69
A0A2H1WXX5
A0A2W1B0I9
A0A212ENY7
S4PTI3
U5EK72
+ More
A0A336MHC0 A0A336M8M2 A0A2M4ACU7 A0A2M4ACN3 A0A2M4BF49 W5JVZ9 A0A182FJW3 A0A2M3ZGY9 A0A2M3ZGV1 B0WWT3 A0A1B0GJC1 A0A182YCS7 A0A2M4BDS4 A0A2M4BDN3 A0A182STD2 A0A1L8DW32 A0A1L8DW35 A0A1S4JVJ2 A0A084VX58 A0A1Q3FW62 A0A023EUD7 A0A182RKY1 A0A1J1IV32 A0A182MF06 A0A182IXL1 A0A1I8MFH2 A0A182H9C6 Q7QFC0 A0A1Y9J009 A0A182K826 Q4V6M6 A0A1I8Q5V5 B4F5R2 A0A0K8TYT0 A0A182QR47 B4F5R3 A0A034WE83 A0A0A1WM66 Q29GE4 B4HB72 B4F5S4 B4IF53 A0A182PT47 A0A182W5T1 A0A0P6J688 A0A3B0KE70 W8C6N5 A0A1A9YGV9 Q179S2 A0A1B0FM02 A0A1A9X010 A0A1B0AJ06 B9EQY8 A0A1B0AT06 A0A1A9UV21 A0A0L0BUP2 A0A1W4WDI3 B4JJD5 B3N0T6 A8JV07 A0A0Q5TK49 A0A182N0K8 A0A0A9XTF3 B4L868 E2AX83 A0A1B6CVX6 B4NCM6 E0VVM6 A0A1B6IAK4 A0A1B6G047 A0A3L8DFW1 D6WHW2 E2BN92 A0A158NVV9 A0A1Y1LHS4 A0A154PII2 A0A1B6L9I5 A0A195ET90 A0A084WR77 A0A0M4EHN9 A0A2R7WAG7 A0A0P4VPV5 A0A087ZTS6 A0A2M4ADQ2 A0A0M8ZTJ4 A0A026WH59 A0A2A3EDY3 A0A3Q0JL82 A0A067QQ33 A0A0J7KCP4 A0A2J7QM59 A0A023F1G4 A0A0V0G744 A0A023F3K2
A0A336MHC0 A0A336M8M2 A0A2M4ACU7 A0A2M4ACN3 A0A2M4BF49 W5JVZ9 A0A182FJW3 A0A2M3ZGY9 A0A2M3ZGV1 B0WWT3 A0A1B0GJC1 A0A182YCS7 A0A2M4BDS4 A0A2M4BDN3 A0A182STD2 A0A1L8DW32 A0A1L8DW35 A0A1S4JVJ2 A0A084VX58 A0A1Q3FW62 A0A023EUD7 A0A182RKY1 A0A1J1IV32 A0A182MF06 A0A182IXL1 A0A1I8MFH2 A0A182H9C6 Q7QFC0 A0A1Y9J009 A0A182K826 Q4V6M6 A0A1I8Q5V5 B4F5R2 A0A0K8TYT0 A0A182QR47 B4F5R3 A0A034WE83 A0A0A1WM66 Q29GE4 B4HB72 B4F5S4 B4IF53 A0A182PT47 A0A182W5T1 A0A0P6J688 A0A3B0KE70 W8C6N5 A0A1A9YGV9 Q179S2 A0A1B0FM02 A0A1A9X010 A0A1B0AJ06 B9EQY8 A0A1B0AT06 A0A1A9UV21 A0A0L0BUP2 A0A1W4WDI3 B4JJD5 B3N0T6 A8JV07 A0A0Q5TK49 A0A182N0K8 A0A0A9XTF3 B4L868 E2AX83 A0A1B6CVX6 B4NCM6 E0VVM6 A0A1B6IAK4 A0A1B6G047 A0A3L8DFW1 D6WHW2 E2BN92 A0A158NVV9 A0A1Y1LHS4 A0A154PII2 A0A1B6L9I5 A0A195ET90 A0A084WR77 A0A0M4EHN9 A0A2R7WAG7 A0A0P4VPV5 A0A087ZTS6 A0A2M4ADQ2 A0A0M8ZTJ4 A0A026WH59 A0A2A3EDY3 A0A3Q0JL82 A0A067QQ33 A0A0J7KCP4 A0A2J7QM59 A0A023F1G4 A0A0V0G744 A0A023F3K2
PDB
3NFQ
E-value=2.90584e-10,
Score=156
Ontologies
GO
Topology
Subcellular location
Nucleus
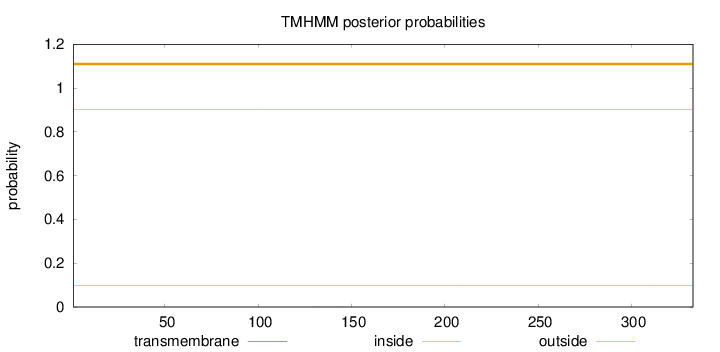
Length:
333
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00506
Exp number, first 60 AAs:
0
Total prob of N-in:
0.09823
outside
1 - 333
Population Genetic Test Statistics
Pi
155.754485
Theta
144.998437
Tajima's D
0.433955
CLR
0
CSRT
0.494975251237438
Interpretation
Uncertain
