Gene
KWMTBOMO14699 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009571
Annotation
Receptor-interacting_serine/threonine-protein_kinase_4_[Papilio_machaon]
Full name
Histone-lysine N-methyltransferase EHMT1
Alternative Name
Euchromatic histone-lysine N-methyltransferase 1
G9a-like protein 1
Lysine N-methyltransferase 1D
G9a-like protein 1
Lysine N-methyltransferase 1D
Location in the cell
Cytoplasmic Reliability : 1.335 Extracellular Reliability : 1.02 Nuclear Reliability : 1.56
Sequence
CDS
ATGTGTTCCGAAGCAGAGAAAGCTCAGTTGTTTGAAGCTATCGACAACAATAATGTAGAAAAAGCTGACGTACTTCTATCTTCAACTGTGGACGTTAACGTACGGAGTCTCGAAGTGAAGAATCGTACTGCGCTGCACCTGGCAGCCCACGCGGGGCGCTACGACCTAGTCGCTCTGCTAGTCGACAAACACAAAGCAGACGTCAACATCGAAGACAGCGAAGGCGACATCCCCTTGCAGCTAGCCACAGATCGCCATCATATGAAGATAGTCAAGTTCCTCCTAAGCAGAAACTCCAAAGGGCGTGAGTACGCGGCGCATTTCCTAAACACTGACGAAGTAGAGGAATTCTTCCAAGCGGCCAAAGCGAATGACATCCAGAGAGTGACGGAGCTACTCGACAGTGGTGTGGATGTCAATTCGGTAGATTTGTCTGATCTACACCGATCCGCGTTGCATGTTGCAGCTCGGGAAGGTCATTATGATCTAGTGCAACTTCTCCTGGAGCGTGGTGCTGATATGCAATACGAAGACGACACTGATGAGTGCGCCTACCACATAGCCCTTAATAACGACCACGACAATATAGGCAGTTTGCTCTTAAAAACAGGGTATATTCCCGACCCGCGCGACGACTCCAGTGTAAGCAGTGACACTGATGAAAGTGTAAGCTTAAGCTTTCTTTTTAACTAA
Protein
MCSEAEKAQLFEAIDNNNVEKADVLLSSTVDVNVRSLEVKNRTALHLAAHAGRYDLVALLVDKHKADVNIEDSEGDIPLQLATDRHHMKIVKFLLSRNSKGREYAAHFLNTDEVEEFFQAAKANDIQRVTELLDSGVDVNSVDLSDLHRSALHVAAREGHYDLVQLLLERGADMQYEDDTDECAYHIALNNDHDNIGSLLLKTGYIPDPRDDSSVSSDTDESVSLSFLFN
Summary
Description
Histone methyltransferase that specifically mono- and dimethylates 'Lys-9' of histone H3 (H3K9me1 and H3K9me2, respectively) in euchromatin. H3K9me represents a specific tag for epigenetic transcriptional repression by recruiting HP1 proteins to methylated histones. Also weakly methylates 'Lys-27' of histone H3 (H3K27me). Also required for DNA methylation, the histone methyltransferase activity is not required for DNA methylation, suggesting that these 2 activities function independently. Probably targeted to histone H3 by different DNA-binding proteins like E2F6, MGA, MAX and/or DP1. During G0 phase, it probably contributes to silencing of MYC- and E2F-responsive genes, suggesting a role in G0/G1 transition in cell cycle. In addition to the histone methyltransferase activity, also methylates non-histone proteins: mediates dimethylation of 'Lys-373' of p53/TP53.
Catalytic Activity
L-lysyl-[histone] + S-adenosyl-L-methionine = H(+) + N(6)-methyl-L-lysyl-[histone] + S-adenosyl-L-homocysteine
Subunit
Interacts with WIZ. Part of the E2F6.com-1 complex in G0 phase composed of E2F6, MGA, MAX, TFDP1, CBX3, BAT8, EHMT1, RING1, RNF2, MBLR, L3MBTL2 and YAF2. Interacts with MPHOSPH8 (By similarity). Interacts with CDYL. Interacts with REST only in the presence of CDYL. Part of a complex containing at least CDYL, REST, WIZ, SETB1, EHMT1 and EHMT2 (By similarity). Heterodimer; heterodimerizes with EHMT2. Interacts (via ANK repeats) with RELA (when monomethylated at 'Lys-310').
Similarity
Belongs to the class V-like SAM-binding methyltransferase superfamily.
Keywords
Acetylation
Alternative splicing
ANK repeat
Chromatin regulator
Chromosome
Complete proteome
Isopeptide bond
Metal-binding
Methyltransferase
Nucleus
Phosphoprotein
Reference proteome
Repeat
S-adenosyl-L-methionine
Transferase
Ubl conjugation
Zinc
Feature
chain Histone-lysine N-methyltransferase EHMT1
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9JJ70
A0A194RW00
A0A194Q060
A0A154NY84
A0A090MA93
A0A087V491
+ More
B3EU24 C3NIT9 X1H8V7 A0A1F9AH59 A0A2P6KIZ5 A0A0P7V8U0 A0A3M0IXE2 U3JSN1 I4BZW7 A0A0B8N264 A0A091SQI4 A0A1W4YJ43 A0A225V0M0 H0YXA8 A0A1S3GA49 W5M0N2 A0A218UDV4 A0A1S3G7V3 A0A1S3G885 A0A2S7Q6R5 A0A093SJC5 U3IXU8 A0A3B0YXP3 G2Y6T1 A0A384J8K7 M7TUB6 A0A091IZR3 A0A091QV00 A0A3Q0H4H9 A0A1V4J783 A0A287DCB6 R0L5Y1 A0A3Q0H944 I3LWJ6 A0A091CQK6 A0A1V4J7B8 A0A2K6GI19 Q47DZ9 A0A2J8RMH6 Q4EC44 L8HI15 Q8BHB2 A0A1V3FVR2 A0A2J8PWU5 A0A1B0GWF6 F6UJH7 A0A1D5PVY2 A0A3Q0H4J8 G3UQS6 G3WHW7 A0A3Q0H949 A0A3Q0H964 A0A3L7HD24 A0A3Q0H4I9 A0A1D5PW08 G5BG04 C0R391 Z4YJZ7 A0A1D5PJ82 A0A151MF60 Q5DW34-2 A0A1U7SD70 A0A0H2UH19 A0A250Y7B8 A0A151MF43 Q5DW34 K7FDM7 G3I5Z2 A0A151MFI4 A0A0P6J5K3 A0A3L8SDY2 H9GIS7 K7FDN4 Q73I01 A0A3Q2U3H4 A0A226PMU1 A0A1D5PIX7 A0A3Q0H5S0 A0A151MF76 A0A0Q3MA63 E9Q5A3 H2R5Y8 D4A005 A0A226NG00 A0A1A6GSP9 H0V8M4 H7CHP8 G1RDJ8 Q5DW34-3 G1MU25 A0A286XW35 A0A1D5P7T6 Q5F3H1 A0A1D5P0G7
B3EU24 C3NIT9 X1H8V7 A0A1F9AH59 A0A2P6KIZ5 A0A0P7V8U0 A0A3M0IXE2 U3JSN1 I4BZW7 A0A0B8N264 A0A091SQI4 A0A1W4YJ43 A0A225V0M0 H0YXA8 A0A1S3GA49 W5M0N2 A0A218UDV4 A0A1S3G7V3 A0A1S3G885 A0A2S7Q6R5 A0A093SJC5 U3IXU8 A0A3B0YXP3 G2Y6T1 A0A384J8K7 M7TUB6 A0A091IZR3 A0A091QV00 A0A3Q0H4H9 A0A1V4J783 A0A287DCB6 R0L5Y1 A0A3Q0H944 I3LWJ6 A0A091CQK6 A0A1V4J7B8 A0A2K6GI19 Q47DZ9 A0A2J8RMH6 Q4EC44 L8HI15 Q8BHB2 A0A1V3FVR2 A0A2J8PWU5 A0A1B0GWF6 F6UJH7 A0A1D5PVY2 A0A3Q0H4J8 G3UQS6 G3WHW7 A0A3Q0H949 A0A3Q0H964 A0A3L7HD24 A0A3Q0H4I9 A0A1D5PW08 G5BG04 C0R391 Z4YJZ7 A0A1D5PJ82 A0A151MF60 Q5DW34-2 A0A1U7SD70 A0A0H2UH19 A0A250Y7B8 A0A151MF43 Q5DW34 K7FDM7 G3I5Z2 A0A151MFI4 A0A0P6J5K3 A0A3L8SDY2 H9GIS7 K7FDN4 Q73I01 A0A3Q2U3H4 A0A226PMU1 A0A1D5PIX7 A0A3Q0H5S0 A0A151MF76 A0A0Q3MA63 E9Q5A3 H2R5Y8 D4A005 A0A226NG00 A0A1A6GSP9 H0V8M4 H7CHP8 G1RDJ8 Q5DW34-3 G1MU25 A0A286XW35 A0A1D5P7T6 Q5F3H1 A0A1D5P0G7
Pubmed
19121390
26354079
20023027
19435847
24624126
27774985
+ More
20360741 23749191 21876677 23104368 26913498 23704180 19650930 15774024 23375108 10349636 11042159 11076861 11217851 12466851 16141073 15164053 21406692 23186163 25755297 28112733 17495919 15592404 20838655 21709235 29704459 21993625 19307581 19468303 22293439 15774718 15489334 18818694 21131967 28087693 17381049 21804562 30282656 21881562 15024419 24621616 16136131 15057822 21993624 15642098
20360741 23749191 21876677 23104368 26913498 23704180 19650930 15774024 23375108 10349636 11042159 11076861 11217851 12466851 16141073 15164053 21406692 23186163 25755297 28112733 17495919 15592404 20838655 21709235 29704459 21993625 19307581 19468303 22293439 15774718 15489334 18818694 21131967 28087693 17381049 21804562 30282656 21881562 15024419 24621616 16136131 15057822 21993624 15642098
EMBL
BABH01036440
KQ459700
KPJ20366.1
KQ459582
KPI98946.1
KQ434782
+ More
KZC04541.1 CBMG010003847 CEG04029.1 KL479275 KFO07433.1 CP001102 ACE06726.1 CP001404 ACP49049.1 BARU01029347 GAH65832.1 MGQK01000096 OGP57425.1 MWRG01010813 PRD26298.1 JARO02002970 KPP71466.1 QRBI01000209 RMB93514.1 AGTO01021651 CP003360 AFM22858.1 DF933843 GAM43281.1 KK931406 KFQ45237.1 NBNE01009010 OWY98851.1 ABQF01048574 ABQF01048575 ABQF01048576 ABQF01048577 ABQF01048578 ABQF01048579 ABQF01048580 AHAT01021783 MUZQ01000386 OWK51903.1 NJPR01000087 PQE22229.1 KL671500 KFW82784.1 ADON01124535 UOFK01000175 VAW78959.1 FQ790293 CCD48333.1 CP009806 ATZ46664.1 KB707926 EMR84885.1 KL218434 KFP05071.1 KK804840 KFQ31380.1 LSYS01008925 OPJ67914.1 AGTP01090670 AGTP01090671 KB744204 EOA95687.1 KN124375 KFO21439.1 OPJ67915.1 CP000089 AAZ46932.1 NDHI03003669 PNJ09655.1 AAGB01000025 EAL59018.1 KB007840 ELR24041.1 AK049454 AK052174 AK082062 BAC33756.1 BAC34869.1 BAC38402.1 MQAD01000002 OOE05664.1 NBAG03000141 PNI88490.1 AL365502 AL590627 AL611925 AADN05000150 AEFK01023222 AEFK01023223 AEFK01023224 AEFK01023225 AEFK01023226 AEFK01023227 AEFK01023228 AEFK01023229 AEFK01023230 AEFK01023231 RAZU01000235 RLQ63844.1 JH170063 EHB08215.1 CP001391 ACN95383.1 AL732525 AKHW03006215 KYO23149.1 AB205007 BC056938 BC089302 GFFV01000418 JAV39527.1 KYO23148.1 AGCU01190420 AGCU01190421 AGCU01190422 AGCU01190423 AGCU01190424 AGCU01190425 AGCU01190426 AGCU01190427 AGCU01190428 AGCU01190429 JH001326 EGW07293.1 KYO23150.1 GEBF01005764 JAN97868.1 QUSF01000027 RLW00329.1 AAWZ02030207 AAWZ02030208 AAWZ02030209 AE017196 AAS14111.1 AWGT02000046 OXB80677.1 KYO23151.1 LMAW01002595 KQK79328.1 AACZ04062180 AACZ04062181 AACZ04062182 AACZ04062183 AACZ04062184 AACZ04062185 AABR07051230 AABR07051231 AABR07051232 AABR07051233 AABR07051234 AABR07051235 AABR07051236 MCFN01000062 OXB66474.1 LZPO01075853 OBS68944.1 AAKN02024151 AAKN02024152 AAKN02024153 AAKN02024154 AAKN02024155 AB702942 BAL72057.1 ADFV01120965 ADFV01120966 ADFV01120967 ADFV01120968 ADFV01120969 ADFV01120970 ADFV01120971 ADFV01120972 ADFV01120973 ADFV01120974 AJ851679 CAH65313.1
KZC04541.1 CBMG010003847 CEG04029.1 KL479275 KFO07433.1 CP001102 ACE06726.1 CP001404 ACP49049.1 BARU01029347 GAH65832.1 MGQK01000096 OGP57425.1 MWRG01010813 PRD26298.1 JARO02002970 KPP71466.1 QRBI01000209 RMB93514.1 AGTO01021651 CP003360 AFM22858.1 DF933843 GAM43281.1 KK931406 KFQ45237.1 NBNE01009010 OWY98851.1 ABQF01048574 ABQF01048575 ABQF01048576 ABQF01048577 ABQF01048578 ABQF01048579 ABQF01048580 AHAT01021783 MUZQ01000386 OWK51903.1 NJPR01000087 PQE22229.1 KL671500 KFW82784.1 ADON01124535 UOFK01000175 VAW78959.1 FQ790293 CCD48333.1 CP009806 ATZ46664.1 KB707926 EMR84885.1 KL218434 KFP05071.1 KK804840 KFQ31380.1 LSYS01008925 OPJ67914.1 AGTP01090670 AGTP01090671 KB744204 EOA95687.1 KN124375 KFO21439.1 OPJ67915.1 CP000089 AAZ46932.1 NDHI03003669 PNJ09655.1 AAGB01000025 EAL59018.1 KB007840 ELR24041.1 AK049454 AK052174 AK082062 BAC33756.1 BAC34869.1 BAC38402.1 MQAD01000002 OOE05664.1 NBAG03000141 PNI88490.1 AL365502 AL590627 AL611925 AADN05000150 AEFK01023222 AEFK01023223 AEFK01023224 AEFK01023225 AEFK01023226 AEFK01023227 AEFK01023228 AEFK01023229 AEFK01023230 AEFK01023231 RAZU01000235 RLQ63844.1 JH170063 EHB08215.1 CP001391 ACN95383.1 AL732525 AKHW03006215 KYO23149.1 AB205007 BC056938 BC089302 GFFV01000418 JAV39527.1 KYO23148.1 AGCU01190420 AGCU01190421 AGCU01190422 AGCU01190423 AGCU01190424 AGCU01190425 AGCU01190426 AGCU01190427 AGCU01190428 AGCU01190429 JH001326 EGW07293.1 KYO23150.1 GEBF01005764 JAN97868.1 QUSF01000027 RLW00329.1 AAWZ02030207 AAWZ02030208 AAWZ02030209 AE017196 AAS14111.1 AWGT02000046 OXB80677.1 KYO23151.1 LMAW01002595 KQK79328.1 AACZ04062180 AACZ04062181 AACZ04062182 AACZ04062183 AACZ04062184 AACZ04062185 AABR07051230 AABR07051231 AABR07051232 AABR07051233 AABR07051234 AABR07051235 AABR07051236 MCFN01000062 OXB66474.1 LZPO01075853 OBS68944.1 AAKN02024151 AAKN02024152 AAKN02024153 AAKN02024154 AAKN02024155 AB702942 BAL72057.1 ADFV01120965 ADFV01120966 ADFV01120967 ADFV01120968 ADFV01120969 ADFV01120970 ADFV01120971 ADFV01120972 ADFV01120973 ADFV01120974 AJ851679 CAH65313.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000076502
UP000001227
UP000006818
+ More
UP000034805 UP000269221 UP000016665 UP000006055 UP000192224 UP000198211 UP000007754 UP000081671 UP000018468 UP000197619 UP000238813 UP000053258 UP000016666 UP000008177 UP000001798 UP000012045 UP000054308 UP000189705 UP000190648 UP000005215 UP000028990 UP000233160 UP000000550 UP000011083 UP000188458 UP000005640 UP000002280 UP000000539 UP000001645 UP000007648 UP000273346 UP000006813 UP000001293 UP000000589 UP000050525 UP000007267 UP000001075 UP000276834 UP000001646 UP000008215 UP000198419 UP000051836 UP000002277 UP000002494 UP000198323 UP000092124 UP000005447 UP000001073
UP000034805 UP000269221 UP000016665 UP000006055 UP000192224 UP000198211 UP000007754 UP000081671 UP000018468 UP000197619 UP000238813 UP000053258 UP000016666 UP000008177 UP000001798 UP000012045 UP000054308 UP000189705 UP000190648 UP000005215 UP000028990 UP000233160 UP000000550 UP000011083 UP000188458 UP000005640 UP000002280 UP000000539 UP000001645 UP000007648 UP000273346 UP000006813 UP000001293 UP000000589 UP000050525 UP000007267 UP000001075 UP000276834 UP000001646 UP000008215 UP000198419 UP000051836 UP000002277 UP000002494 UP000198323 UP000092124 UP000005447 UP000001073
Pfam
Interpro
IPR002110
Ankyrin_rpt
+ More
IPR020683 Ankyrin_rpt-contain_dom
IPR036770 Ankyrin_rpt-contain_sf
IPR002893 Znf_MYND
IPR001214 SET_dom
IPR038035 EHMT1
IPR007728 Pre-SET_dom
IPR013083 Znf_RING/FYVE/PHD
IPR003613 Ubox_domain
IPR036390 WH_DNA-bd_sf
IPR036388 WH-like_DNA-bd_sf
IPR001995 Peptidase_A2_cat
IPR031352 SesA
IPR027417 P-loop_NTPase
IPR020683 Ankyrin_rpt-contain_dom
IPR036770 Ankyrin_rpt-contain_sf
IPR002893 Znf_MYND
IPR001214 SET_dom
IPR038035 EHMT1
IPR007728 Pre-SET_dom
IPR013083 Znf_RING/FYVE/PHD
IPR003613 Ubox_domain
IPR036390 WH_DNA-bd_sf
IPR036388 WH-like_DNA-bd_sf
IPR001995 Peptidase_A2_cat
IPR031352 SesA
IPR027417 P-loop_NTPase
Gene 3D
CDD
ProteinModelPortal
H9JJ70
A0A194RW00
A0A194Q060
A0A154NY84
A0A090MA93
A0A087V491
+ More
B3EU24 C3NIT9 X1H8V7 A0A1F9AH59 A0A2P6KIZ5 A0A0P7V8U0 A0A3M0IXE2 U3JSN1 I4BZW7 A0A0B8N264 A0A091SQI4 A0A1W4YJ43 A0A225V0M0 H0YXA8 A0A1S3GA49 W5M0N2 A0A218UDV4 A0A1S3G7V3 A0A1S3G885 A0A2S7Q6R5 A0A093SJC5 U3IXU8 A0A3B0YXP3 G2Y6T1 A0A384J8K7 M7TUB6 A0A091IZR3 A0A091QV00 A0A3Q0H4H9 A0A1V4J783 A0A287DCB6 R0L5Y1 A0A3Q0H944 I3LWJ6 A0A091CQK6 A0A1V4J7B8 A0A2K6GI19 Q47DZ9 A0A2J8RMH6 Q4EC44 L8HI15 Q8BHB2 A0A1V3FVR2 A0A2J8PWU5 A0A1B0GWF6 F6UJH7 A0A1D5PVY2 A0A3Q0H4J8 G3UQS6 G3WHW7 A0A3Q0H949 A0A3Q0H964 A0A3L7HD24 A0A3Q0H4I9 A0A1D5PW08 G5BG04 C0R391 Z4YJZ7 A0A1D5PJ82 A0A151MF60 Q5DW34-2 A0A1U7SD70 A0A0H2UH19 A0A250Y7B8 A0A151MF43 Q5DW34 K7FDM7 G3I5Z2 A0A151MFI4 A0A0P6J5K3 A0A3L8SDY2 H9GIS7 K7FDN4 Q73I01 A0A3Q2U3H4 A0A226PMU1 A0A1D5PIX7 A0A3Q0H5S0 A0A151MF76 A0A0Q3MA63 E9Q5A3 H2R5Y8 D4A005 A0A226NG00 A0A1A6GSP9 H0V8M4 H7CHP8 G1RDJ8 Q5DW34-3 G1MU25 A0A286XW35 A0A1D5P7T6 Q5F3H1 A0A1D5P0G7
B3EU24 C3NIT9 X1H8V7 A0A1F9AH59 A0A2P6KIZ5 A0A0P7V8U0 A0A3M0IXE2 U3JSN1 I4BZW7 A0A0B8N264 A0A091SQI4 A0A1W4YJ43 A0A225V0M0 H0YXA8 A0A1S3GA49 W5M0N2 A0A218UDV4 A0A1S3G7V3 A0A1S3G885 A0A2S7Q6R5 A0A093SJC5 U3IXU8 A0A3B0YXP3 G2Y6T1 A0A384J8K7 M7TUB6 A0A091IZR3 A0A091QV00 A0A3Q0H4H9 A0A1V4J783 A0A287DCB6 R0L5Y1 A0A3Q0H944 I3LWJ6 A0A091CQK6 A0A1V4J7B8 A0A2K6GI19 Q47DZ9 A0A2J8RMH6 Q4EC44 L8HI15 Q8BHB2 A0A1V3FVR2 A0A2J8PWU5 A0A1B0GWF6 F6UJH7 A0A1D5PVY2 A0A3Q0H4J8 G3UQS6 G3WHW7 A0A3Q0H949 A0A3Q0H964 A0A3L7HD24 A0A3Q0H4I9 A0A1D5PW08 G5BG04 C0R391 Z4YJZ7 A0A1D5PJ82 A0A151MF60 Q5DW34-2 A0A1U7SD70 A0A0H2UH19 A0A250Y7B8 A0A151MF43 Q5DW34 K7FDM7 G3I5Z2 A0A151MFI4 A0A0P6J5K3 A0A3L8SDY2 H9GIS7 K7FDN4 Q73I01 A0A3Q2U3H4 A0A226PMU1 A0A1D5PIX7 A0A3Q0H5S0 A0A151MF76 A0A0Q3MA63 E9Q5A3 H2R5Y8 D4A005 A0A226NG00 A0A1A6GSP9 H0V8M4 H7CHP8 G1RDJ8 Q5DW34-3 G1MU25 A0A286XW35 A0A1D5P7T6 Q5F3H1 A0A1D5P0G7
PDB
6BY9
E-value=1.33971e-16,
Score=208
Ontologies
GO
GO:0016301
GO:0046872
GO:0018024
GO:0005694
GO:0005634
GO:0002039
GO:0008270
GO:0004842
GO:0016021
GO:0046976
GO:0018027
GO:0046974
GO:0070742
GO:0000122
GO:0018026
GO:0045995
GO:0006306
GO:0120162
GO:0016604
GO:0004190
GO:0008168
GO:0006325
GO:0016571
GO:0045892
GO:0005654
GO:0060992
GO:0016279
GO:0005515
GO:0016020
GO:0017038
GO:0006605
GO:0006886
GO:0003824
GO:0008152
GO:0016491
GO:0015930
PANTHER
Topology
Subcellular location
Chromosome
Nucleus
Nucleus
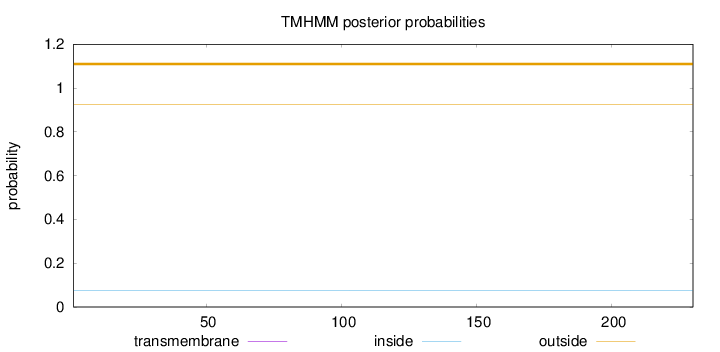
Length:
230
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00036
Exp number, first 60 AAs:
0.00035
Total prob of N-in:
0.07589
outside
1 - 230
Population Genetic Test Statistics
Pi
330.993714
Theta
216.693499
Tajima's D
1.362947
CLR
0
CSRT
0.750762461876906
Interpretation
Uncertain
