Gene
KWMTBOMO14694
Pre Gene Modal
BGIBMGA009620
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_E3_ubiquitin-protein_ligase_RFWD2-like_[Plutella_xylostella]
Full name
E3 ubiquitin-protein ligase COP1
Alternative Name
Constitutive photomorphogenesis protein 1 homolog
RING finger and WD repeat domain protein 2
RING finger protein 200
RING-type E3 ubiquitin transferase RFWD2
RING finger and WD repeat domain protein 2
RING finger protein 200
RING-type E3 ubiquitin transferase RFWD2
Location in the cell
Nuclear Reliability : 1.065
Sequence
CDS
ATGATTCAATTAGAAAGTAGCGCGGGCGCGGTGTGCTCGGAGGAGGAGAGCTTCGCGGGCGGCGCGGGGGGCGGGGGGGACGCCCTGGCCGCGCGCTGGAGGAAGCTCACGCAACACTTCGACGACTTCGTGCAGTGTTACTTCGCGCACCGCGCTGACGAGCTGTACTTCCCGGGGTCGGGGTCGCTGACCCCCGCGTCCCCCGCCGTCCCCGCCTACCCCGCACCCCCCACCGTGACCCCCGTGCCCTCCTCGGCGGCCCCGTACACGTGCGAGGCGGACCACGTGCCGCAGGGTCCGGTGTGCGACCCGCAACCAACTCCTGCACCGGGTCCGTCGGACCAGCGGGCGCCGGTGGCGGGCGTGGCGGTCGAGGACTCCGTGGAGATGGAGGAGGCCGGCTCCACTCGAGCCCGCGCCGGACTCGACGCCTTCCGGGACGACCTGGTCGCTTTCACCGCCTACAGGACGCTACGGCCGCTGGCCACGCTGTCGTACTGCAGCGACGCCATCAACTACTCCACGATCGTGTCCACCATCGAGTTCGACAAGGACGAGGAGTTCTTCGCGATCGCCGGCGTCACCAAGAGGATCAAGGTGTTCGAGTTCGAGTCGGTGGTGCGCGACGCGGTGGACGTGCACTACCCGTGCGCAGAGATGCAGTGCTCGCACAAGATCTCGTGCGTGTCGTGGAACAAGTACCACAAGCACGTGCTGGCGTCGTCGGACTACGAGGGCACGGTGTCGGTGTGGGACGCGGGCACGGCGGTGCGCACGCGCGCGCTGCAGGAGCACGACAAGCGCGCCTGGTCCGTGCACTTCAACCGCGCCGACGTGCGCCTGCTGGCCTCGGGCTCCGACGACGCGCGCGTCAAGCTGTGGGCGCTCAACCAGGAGAAGTCCGTCGCCACGCTCGAGGCCAAGTTCAACGTGTGCTGCGTGCGCTTCAACCCCAACTCGTCCTGCCACCTGGCCTTCGGCTCCGCAGACCACTGCGTGCACTACTACGACCTGCGCTCGCCGCGCGCGCCGCTGGCCGTGTTCCGGGACCACCGCAAGGCGGTCTCGTACGTGAAGTTCCTGGACGGCGGCACGCTGGTGTCCGCCTCCACGGACTCGCAGCTCAAGCTGTGGCGCGTGGAGTCCCCGCGCTGCGTGCGCTCCTTCACCGGACACACCAACGAGAAGAACTTTGTGGGGCTGGCCACCGACGGCCGCTACGTGGCGTGCGGCTCGGAGAACAACGCGCTCTACGTGTACCACAGCGGCCTGGCGCGCCCCTGCCTCGCCTACAGGTTCGACCCCGCGCGCCCCCTCCTCGAGCGCGACCGCCGCGACGAGGAGCCCGCAGAGTTCGTCTCCGCCGTCTGCTGGAGGAGGGCGGCCTCCCGCCCCGCCCTCCTCGCCGCCAACAGCCACGGCACCATCAAGGTCCTCGAGCTCGTCTGA
Protein
MIQLESSAGAVCSEEESFAGGAGGGGDALAARWRKLTQHFDDFVQCYFAHRADELYFPGSGSLTPASPAVPAYPAPPTVTPVPSSAAPYTCEADHVPQGPVCDPQPTPAPGPSDQRAPVAGVAVEDSVEMEEAGSTRARAGLDAFRDDLVAFTAYRTLRPLATLSYCSDAINYSTIVSTIEFDKDEEFFAIAGVTKRIKVFEFESVVRDAVDVHYPCAEMQCSHKISCVSWNKYHKHVLASSDYEGTVSVWDAGTAVRTRALQEHDKRAWSVHFNRADVRLLASGSDDARVKLWALNQEKSVATLEAKFNVCCVRFNPNSSCHLAFGSADHCVHYYDLRSPRAPLAVFRDHRKAVSYVKFLDGGTLVSASTDSQLKLWRVESPRCVRSFTGHTNEKNFVGLATDGRYVACGSENNALYVYHSGLARPCLAYRFDPARPLLERDRRDEEPAEFVSAVCWRRAASRPALLAANSHGTIKVLELV
Summary
Description
E3 ubiquitin-protein ligase that mediates ubiquitination and subsequent proteasomal degradation of target proteins. E3 ubiquitin ligases accept ubiquitin from an E2 ubiquitin-conjugating enzyme in the form of a thioester and then directly transfers the ubiquitin to targeted substrates. Involved in JUN ubiquitination and degradation. Directly involved in p53 (TP53) ubiquitination and degradation, thereby abolishing p53-dependent transcription and apoptosis. Ubiquitinates p53 independently of MDM2 or RCHY1. Probably mediates E3 ubiquitin ligase activity by functioning as the essential RING domain subunit of larger E3 complexes. In contrast, it does not constitute the catalytic RING subunit in the DCX DET1-COP1 complex that negatively regulates JUN, the ubiquitin ligase activity being mediated by RBX1. Involved in 14-3-3 protein sigma/SFN ubiquitination and proteasomal degradation, leading to AKT activation and promotion of cell survival. Ubiquitinates MTA1 leading to its proteasomal degradation. Upon binding to TRIB1, ubiquitinates CEBPA, which lacks a canonical COP1-binding motif (Probable).
Catalytic Activity
S-ubiquitinyl-[E2 ubiquitin-conjugating enzyme]-L-cysteine + [acceptor protein]-L-lysine = [E2 ubiquitin-conjugating enzyme]-L-cysteine + N(6)-ubiquitinyl-[acceptor protein]-L-lysine.
Subunit
Homodimer. Homodimerization is mediated by the coiled coil domain. Component of the DCX DET1-COP1 ubiquitin ligase complex at least composed of RBX1, DET1, DDB1, CUL4A and COP1. Isoform 2 does not interact with CUL4A but still binds to RBX1, suggesting that the interaction may be mediated by another cullin protein. Isoform 1 and isoform 2 interact with CUL5 but not with CUL1, CUL2 not CUL3. Interacts with bZIP transcription factors JUN, JUNB and JUND but not with FOS, ATF2 nor XBP1. Interacts with p53 (TP53). Interacts with COPS6; this interaction stabilizes RFWD2 through reducing its auto-ubiquitination and decelerating its turnover rate. Interacts with SFN; this interaction leads to SFN degradation. Isoform 4 forms heterodimers with isoform 1, preventing its association with DET1. Interacts with p53/TP53 and MTA1. Interacts with TRIB1 (via C-terminus) and TRIB2 (PubMed:20410507, PubMed:27041596).
Similarity
Belongs to the COP1 family.
Keywords
3D-structure
Alternative splicing
Coiled coil
Complete proteome
Cytoplasm
Metal-binding
Nucleus
Reference proteome
Repeat
Transferase
Ubl conjugation
Ubl conjugation pathway
WD repeat
Zinc
Zinc-finger
Feature
chain E3 ubiquitin-protein ligase COP1
splice variant In isoform 3.
splice variant In isoform 3.
Uniprot
H9JJB9
A0A2H1WAK2
A0A194QU24
A0A212FBZ3
A0A2A4J9J5
A0A2W1B778
+ More
A0A1B6L716 A0A1B6CBX0 A0A2P8YKN3 A0A1B6JH28 A0A1B6FDX3 A0A0N7Z8H0 A0A0J7NTT0 A0A195BRB7 A0A2J7PN84 A0A158NZF3 A0A088ADG8 K7J750 A0A0L7RJM9 E2B9F2 A0A232FG18 A0A2A3EDH8 A0A0M8ZZF6 T1I8V9 A0A154P1Z2 A0A0V0G806 D6WAL1 A0A224XJS6 A0A0A9WSN1 A0A310SJF4 A0A2R7X0M9 A0A0T6BAI2 A0A146LYB2 A0A067R1R4 A0A1Z5LHR7 V5HY20 A0A2R5LMI9 A0A293L690 E0VY83 A0A1Y1MGZ3 A0A131ZAM6 A0A224YWI5 A0A224YXC8 A0A131YQH5 L7M554 A0A131XC23 X1XB79 A0A2H8TLK8 A0A2S2PLB0 A0A026WCC7 N6U1H1 A0A1E1XD65 A0A1A8AFQ6 T1JKE0 Q5BJY0 A0A1U8BR60 A0A340WX38 V9KJ35 A0A250Y1X6 A0A340WPI1 A0A2K5PH54 U6D0K2 A0A1S3WE28 A0A3Q0DY25 A0A340WWG4 A0A2U3XGE1 A0A2U3ZDH9 H9F8U6 A0A2U4CRD5 Q8NHY2-3 A0A1V9XYE5 G3HA05 A0A218V488 A0A2I0MC37 A0A2I0MC58 K7FZ14 A0A2J8N6Z8 A0A2J8UC33 H0Y339 A0A3Q0DU09 A0A1S3JBR8 A0A1U7RGS4 A0A091DK72 A0A3Q0DX67 A0A3Q0DQN3 A0A2U3XGG5 A0A3Q0DQN9 X5D9B8 A0A060WTQ7 A0A1S3WET0 A0A3Q2CY50 U3K105 A0A3Q2CY30 A0A2U4CR21
A0A1B6L716 A0A1B6CBX0 A0A2P8YKN3 A0A1B6JH28 A0A1B6FDX3 A0A0N7Z8H0 A0A0J7NTT0 A0A195BRB7 A0A2J7PN84 A0A158NZF3 A0A088ADG8 K7J750 A0A0L7RJM9 E2B9F2 A0A232FG18 A0A2A3EDH8 A0A0M8ZZF6 T1I8V9 A0A154P1Z2 A0A0V0G806 D6WAL1 A0A224XJS6 A0A0A9WSN1 A0A310SJF4 A0A2R7X0M9 A0A0T6BAI2 A0A146LYB2 A0A067R1R4 A0A1Z5LHR7 V5HY20 A0A2R5LMI9 A0A293L690 E0VY83 A0A1Y1MGZ3 A0A131ZAM6 A0A224YWI5 A0A224YXC8 A0A131YQH5 L7M554 A0A131XC23 X1XB79 A0A2H8TLK8 A0A2S2PLB0 A0A026WCC7 N6U1H1 A0A1E1XD65 A0A1A8AFQ6 T1JKE0 Q5BJY0 A0A1U8BR60 A0A340WX38 V9KJ35 A0A250Y1X6 A0A340WPI1 A0A2K5PH54 U6D0K2 A0A1S3WE28 A0A3Q0DY25 A0A340WWG4 A0A2U3XGE1 A0A2U3ZDH9 H9F8U6 A0A2U4CRD5 Q8NHY2-3 A0A1V9XYE5 G3HA05 A0A218V488 A0A2I0MC37 A0A2I0MC58 K7FZ14 A0A2J8N6Z8 A0A2J8UC33 H0Y339 A0A3Q0DU09 A0A1S3JBR8 A0A1U7RGS4 A0A091DK72 A0A3Q0DX67 A0A3Q0DQN3 A0A2U3XGG5 A0A3Q0DQN9 X5D9B8 A0A060WTQ7 A0A1S3WET0 A0A3Q2CY50 U3K105 A0A3Q2CY30 A0A2U4CR21
EC Number
2.3.2.27
Pubmed
19121390
26354079
22118469
28756777
29403074
27129103
+ More
21347285 20075255 20798317 28648823 18362917 19820115 25401762 26823975 24845553 28528879 25765539 20566863 28004739 26830274 28797301 25576852 28049606 24508170 23537049 28503490 15489334 24402279 28087693 25319552 12466024 12615916 14739464 24722188 14702039 16710414 15103385 17968316 19837670 19805145 20410507 21625211 27041596 28327890 21804562 23371554 17381049 24755649
21347285 20075255 20798317 28648823 18362917 19820115 25401762 26823975 24845553 28528879 25765539 20566863 28004739 26830274 28797301 25576852 28049606 24508170 23537049 28503490 15489334 24402279 28087693 25319552 12466024 12615916 14739464 24722188 14702039 16710414 15103385 17968316 19837670 19805145 20410507 21625211 27041596 28327890 21804562 23371554 17381049 24755649
EMBL
BABH01036433
BABH01036434
BABH01036435
ODYU01007377
SOQ50083.1
KQ461108
+ More
KPJ08972.1 AGBW02009260 OWR51243.1 NWSH01002382 PCG68426.1 KZ150253 PZC71799.1 GEBQ01020465 JAT19512.1 GEDC01026357 JAS10941.1 PYGN01000528 PSN44774.1 GECU01009202 JAS98504.1 GECZ01021343 JAS48426.1 GDKW01003371 JAI53224.1 LBMM01001743 KMQ95825.1 KQ976423 KYM88754.1 NEVH01023957 PNF17804.1 ADTU01000886 AAZX01008222 KQ414581 KOC71003.1 GL446502 EFN87676.1 NNAY01000297 OXU29379.1 KZ288271 PBC29833.1 KQ435812 KOX72753.1 ACPB03024616 KQ434803 KZC05946.1 GECL01001995 JAP04129.1 KQ971312 EEZ97982.1 GFTR01007584 JAW08842.1 GBHO01033174 GBHO01033173 GDHC01008730 JAG10430.1 JAG10431.1 JAQ09899.1 KQ760549 OAD59875.1 KK856260 PTY25326.1 LJIG01002847 KRT84093.1 GDHC01007003 JAQ11626.1 KK852811 KDR15937.1 GFJQ02000055 JAW06915.1 GANP01003086 JAB81382.1 GGLE01006573 MBY10699.1 GFWV01002787 MAA27517.1 DS235845 EEB18339.1 GEZM01031084 GEZM01031083 JAV85072.1 GEDV01000687 JAP87870.1 GFPF01007507 MAA18653.1 GFPF01007458 MAA18604.1 GEDV01007370 JAP81187.1 GACK01006815 JAA58219.1 GEFH01004936 JAP63645.1 ABLF02034147 GFXV01003218 MBW15023.1 GGMR01017439 MBY30058.1 KK107321 EZA52694.1 APGK01053202 KB741222 KB631665 ENN72402.1 ERL85146.1 GFAC01001996 JAT97192.1 HADY01014585 SBP53070.1 AFFK01019697 BC091284 AAH91284.1 JW865412 AFO97929.1 GFFV01002359 JAV37586.1 HAAF01003812 CCP75637.1 JU327299 AFE71055.1 AF508940 BK000438 AF527539 AY509921 KJ534928 KJ535076 AK025789 AK314750 AL162736 AL359265 AL513329 AL590723 CH471067 BC094728 MNPL01002182 OQR78480.1 JH000242 EGV96157.1 MUZQ01000049 OWK60895.1 AKCR02000021 PKK27247.1 PKK27246.1 AGCU01203946 AGCU01203947 AGCU01203948 AGCU01203949 AGCU01203950 NBAG03000235 PNI67549.1 NDHI03003464 PNJ42804.1 KN122390 KFO30665.1 KJ534933 AHW56573.1 FR904722 CDQ70546.1 AGTO01011210
KPJ08972.1 AGBW02009260 OWR51243.1 NWSH01002382 PCG68426.1 KZ150253 PZC71799.1 GEBQ01020465 JAT19512.1 GEDC01026357 JAS10941.1 PYGN01000528 PSN44774.1 GECU01009202 JAS98504.1 GECZ01021343 JAS48426.1 GDKW01003371 JAI53224.1 LBMM01001743 KMQ95825.1 KQ976423 KYM88754.1 NEVH01023957 PNF17804.1 ADTU01000886 AAZX01008222 KQ414581 KOC71003.1 GL446502 EFN87676.1 NNAY01000297 OXU29379.1 KZ288271 PBC29833.1 KQ435812 KOX72753.1 ACPB03024616 KQ434803 KZC05946.1 GECL01001995 JAP04129.1 KQ971312 EEZ97982.1 GFTR01007584 JAW08842.1 GBHO01033174 GBHO01033173 GDHC01008730 JAG10430.1 JAG10431.1 JAQ09899.1 KQ760549 OAD59875.1 KK856260 PTY25326.1 LJIG01002847 KRT84093.1 GDHC01007003 JAQ11626.1 KK852811 KDR15937.1 GFJQ02000055 JAW06915.1 GANP01003086 JAB81382.1 GGLE01006573 MBY10699.1 GFWV01002787 MAA27517.1 DS235845 EEB18339.1 GEZM01031084 GEZM01031083 JAV85072.1 GEDV01000687 JAP87870.1 GFPF01007507 MAA18653.1 GFPF01007458 MAA18604.1 GEDV01007370 JAP81187.1 GACK01006815 JAA58219.1 GEFH01004936 JAP63645.1 ABLF02034147 GFXV01003218 MBW15023.1 GGMR01017439 MBY30058.1 KK107321 EZA52694.1 APGK01053202 KB741222 KB631665 ENN72402.1 ERL85146.1 GFAC01001996 JAT97192.1 HADY01014585 SBP53070.1 AFFK01019697 BC091284 AAH91284.1 JW865412 AFO97929.1 GFFV01002359 JAV37586.1 HAAF01003812 CCP75637.1 JU327299 AFE71055.1 AF508940 BK000438 AF527539 AY509921 KJ534928 KJ535076 AK025789 AK314750 AL162736 AL359265 AL513329 AL590723 CH471067 BC094728 MNPL01002182 OQR78480.1 JH000242 EGV96157.1 MUZQ01000049 OWK60895.1 AKCR02000021 PKK27247.1 PKK27246.1 AGCU01203946 AGCU01203947 AGCU01203948 AGCU01203949 AGCU01203950 NBAG03000235 PNI67549.1 NDHI03003464 PNJ42804.1 KN122390 KFO30665.1 KJ534933 AHW56573.1 FR904722 CDQ70546.1 AGTO01011210
Proteomes
UP000005204
UP000053240
UP000007151
UP000218220
UP000245037
UP000036403
+ More
UP000078540 UP000235965 UP000005205 UP000005203 UP000002358 UP000053825 UP000008237 UP000215335 UP000242457 UP000053105 UP000015103 UP000076502 UP000007266 UP000027135 UP000009046 UP000007819 UP000053097 UP000019118 UP000030742 UP000189706 UP000265300 UP000233040 UP000079721 UP000189704 UP000245341 UP000245340 UP000245320 UP000005640 UP000192247 UP000001075 UP000197619 UP000053872 UP000007267 UP000085678 UP000189705 UP000028990 UP000193380 UP000265020 UP000016665
UP000078540 UP000235965 UP000005205 UP000005203 UP000002358 UP000053825 UP000008237 UP000215335 UP000242457 UP000053105 UP000015103 UP000076502 UP000007266 UP000027135 UP000009046 UP000007819 UP000053097 UP000019118 UP000030742 UP000189706 UP000265300 UP000233040 UP000079721 UP000189704 UP000245341 UP000245340 UP000245320 UP000005640 UP000192247 UP000001075 UP000197619 UP000053872 UP000007267 UP000085678 UP000189705 UP000028990 UP000193380 UP000265020 UP000016665
Interpro
IPR036322
WD40_repeat_dom_sf
+ More
IPR001841 Znf_RING
IPR001680 WD40_repeat
IPR017986 WD40_repeat_dom
IPR013083 Znf_RING/FYVE/PHD
IPR017907 Znf_RING_CS
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR020472 G-protein_beta_WD-40_rep
IPR019775 WD40_repeat_CS
IPR003613 Ubox_domain
IPR008283 Peptidase_M17_N
IPR011356 Leucine_aapep/pepB
IPR000819 Peptidase_M17_C
IPR001841 Znf_RING
IPR001680 WD40_repeat
IPR017986 WD40_repeat_dom
IPR013083 Znf_RING/FYVE/PHD
IPR017907 Znf_RING_CS
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR020472 G-protein_beta_WD-40_rep
IPR019775 WD40_repeat_CS
IPR003613 Ubox_domain
IPR008283 Peptidase_M17_N
IPR011356 Leucine_aapep/pepB
IPR000819 Peptidase_M17_C
SUPFAM
SSF50978
SSF50978
Gene 3D
ProteinModelPortal
H9JJB9
A0A2H1WAK2
A0A194QU24
A0A212FBZ3
A0A2A4J9J5
A0A2W1B778
+ More
A0A1B6L716 A0A1B6CBX0 A0A2P8YKN3 A0A1B6JH28 A0A1B6FDX3 A0A0N7Z8H0 A0A0J7NTT0 A0A195BRB7 A0A2J7PN84 A0A158NZF3 A0A088ADG8 K7J750 A0A0L7RJM9 E2B9F2 A0A232FG18 A0A2A3EDH8 A0A0M8ZZF6 T1I8V9 A0A154P1Z2 A0A0V0G806 D6WAL1 A0A224XJS6 A0A0A9WSN1 A0A310SJF4 A0A2R7X0M9 A0A0T6BAI2 A0A146LYB2 A0A067R1R4 A0A1Z5LHR7 V5HY20 A0A2R5LMI9 A0A293L690 E0VY83 A0A1Y1MGZ3 A0A131ZAM6 A0A224YWI5 A0A224YXC8 A0A131YQH5 L7M554 A0A131XC23 X1XB79 A0A2H8TLK8 A0A2S2PLB0 A0A026WCC7 N6U1H1 A0A1E1XD65 A0A1A8AFQ6 T1JKE0 Q5BJY0 A0A1U8BR60 A0A340WX38 V9KJ35 A0A250Y1X6 A0A340WPI1 A0A2K5PH54 U6D0K2 A0A1S3WE28 A0A3Q0DY25 A0A340WWG4 A0A2U3XGE1 A0A2U3ZDH9 H9F8U6 A0A2U4CRD5 Q8NHY2-3 A0A1V9XYE5 G3HA05 A0A218V488 A0A2I0MC37 A0A2I0MC58 K7FZ14 A0A2J8N6Z8 A0A2J8UC33 H0Y339 A0A3Q0DU09 A0A1S3JBR8 A0A1U7RGS4 A0A091DK72 A0A3Q0DX67 A0A3Q0DQN3 A0A2U3XGG5 A0A3Q0DQN9 X5D9B8 A0A060WTQ7 A0A1S3WET0 A0A3Q2CY50 U3K105 A0A3Q2CY30 A0A2U4CR21
A0A1B6L716 A0A1B6CBX0 A0A2P8YKN3 A0A1B6JH28 A0A1B6FDX3 A0A0N7Z8H0 A0A0J7NTT0 A0A195BRB7 A0A2J7PN84 A0A158NZF3 A0A088ADG8 K7J750 A0A0L7RJM9 E2B9F2 A0A232FG18 A0A2A3EDH8 A0A0M8ZZF6 T1I8V9 A0A154P1Z2 A0A0V0G806 D6WAL1 A0A224XJS6 A0A0A9WSN1 A0A310SJF4 A0A2R7X0M9 A0A0T6BAI2 A0A146LYB2 A0A067R1R4 A0A1Z5LHR7 V5HY20 A0A2R5LMI9 A0A293L690 E0VY83 A0A1Y1MGZ3 A0A131ZAM6 A0A224YWI5 A0A224YXC8 A0A131YQH5 L7M554 A0A131XC23 X1XB79 A0A2H8TLK8 A0A2S2PLB0 A0A026WCC7 N6U1H1 A0A1E1XD65 A0A1A8AFQ6 T1JKE0 Q5BJY0 A0A1U8BR60 A0A340WX38 V9KJ35 A0A250Y1X6 A0A340WPI1 A0A2K5PH54 U6D0K2 A0A1S3WE28 A0A3Q0DY25 A0A340WWG4 A0A2U3XGE1 A0A2U3ZDH9 H9F8U6 A0A2U4CRD5 Q8NHY2-3 A0A1V9XYE5 G3HA05 A0A218V488 A0A2I0MC37 A0A2I0MC58 K7FZ14 A0A2J8N6Z8 A0A2J8UC33 H0Y339 A0A3Q0DU09 A0A1S3JBR8 A0A1U7RGS4 A0A091DK72 A0A3Q0DX67 A0A3Q0DQN3 A0A2U3XGG5 A0A3Q0DQN9 X5D9B8 A0A060WTQ7 A0A1S3WET0 A0A3Q2CY50 U3K105 A0A3Q2CY30 A0A2U4CR21
PDB
5HQG
E-value=8.58299e-119,
Score=1093
Ontologies
GO
GO:0016874
GO:0004842
GO:0032440
GO:0004177
GO:0008235
GO:0005737
GO:0030145
GO:0000139
GO:0005829
GO:0005654
GO:0046872
GO:0043687
GO:0010212
GO:0043161
GO:0031464
GO:0016607
GO:0032436
GO:0061630
GO:0005515
GO:0008270
GO:0042803
GO:0051087
GO:0005634
GO:0006303
GO:0003824
GO:0008152
GO:0016491
GO:0015930
Topology
Subcellular location
Nucleus speckle
Cytoplasm
Cytoplasm
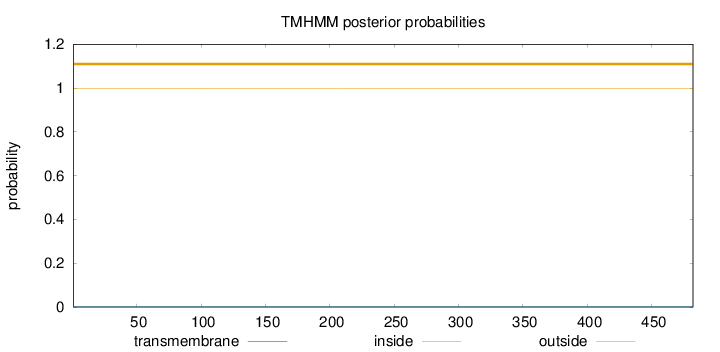
Length:
482
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00133
Exp number, first 60 AAs:
0.00011
Total prob of N-in:
0.00121
outside
1 - 482
Population Genetic Test Statistics
Pi
193.351058
Theta
135.554876
Tajima's D
1.473201
CLR
0
CSRT
0.774061296935153
Interpretation
Uncertain
