Gene
KWMTBOMO14693
Pre Gene Modal
BGIBMGA009620
Annotation
PREDICTED:_E3_ubiquitin-protein_ligase_RFWD2-like_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.623
Sequence
CDS
ATGTCGGGTGCCGCAGCCAGCAGTGCAGGCTCTGGCACGGCCACCACTCTCGGAGGACTTGGCGGCTCACTTGAGGATAAGGACTGTGAGCTCCTGTGCCCGGTGTGCTTCGAGCTGATCGACGAGGCCTATGTGACGCGCTGCGGCCACTCGTTCTGCTACGCGTGCATCAGCCGCGCGCTCGACCTGCACCGGCGCTGTCCCAAGTGCGGAGCGCCCCTGCTCACCAGAGAACACATCTTCCCCAACTTCCTGCTCAATGAACTCGTGACGAAGCGCCGGCTCCGGTCGAGGGCGTCGTCCCGGTCGGGCCCGGCCGGCGAGGGAGCCCGGCTGCGCTCGCTGCTCGCCGCGGAGTCGCGACACCTCGACCTCACCGAGGTGGATTCCATGTTGGAACTGCTCAACAGACGGAAGAGACTGCTGGAGGCGGAGTGCGCGGCGGCGCAGCACCGGCTACTGTACGAGTTCCTGGCGCACCTGCACCGCCACCGCACCCACCAGCTGCAGCAGCTCGCCAGGGAGGCCGCCCTCGTGCACAAGGACATGGAGTACGTCGCCGGAGTGCTGAAGGAACTCCGCACGGGTGCAGTGGGGATAGCGCGCGGGGCCTCGCCCCCCGGCGGCGCGGACCCCCGCCCAGACGCCGTCACTGCGCTCAGGAAAGAGATCGCCGATATAGCCGGGAACCTCGCAGACACGGCCAGCAATGTTGACGGTATTCATTGTTACAATGGTAGTTTTCCAATTACAGCACTATAG
Protein
MSGAAASSAGSGTATTLGGLGGSLEDKDCELLCPVCFELIDEAYVTRCGHSFCYACISRALDLHRRCPKCGAPLLTREHIFPNFLLNELVTKRRLRSRASSRSGPAGEGARLRSLLAAESRHLDLTEVDSMLELLNRRKRLLEAECAAAQHRLLYEFLAHLHRHRTHQLQQLAREAALVHKDMEYVAGVLKELRTGAVGIARGASPPGGADPRPDAVTALRKEIADIAGNLADTASNVDGIHCYNGSFPITAL
Summary
Uniprot
H9JJB9
A0A194QU24
A0A194Q9M7
A0A212FBW2
U5EVR0
W5J2X7
+ More
A0A2P8YKN3 K7J750 A0A1B6CBX0 A0A1B0D6S5 A0A182FSV7 A0A182LAH1 A0A3Q0J4I9 A0A182W135 A0A084W778 F5HJD2 A0A182RRT5 A0A1I8JVR1 A0A182M5P2 A0A182JFK9 A0A182SVX4 A0A1B6FDX3 A0A182I8L7 A0A0M8ZZF6 A0A0P6K128 A0A182K6A7 A0A154P1Z2 A0A182H264 A0A232FG18 T1JKE0 A0A240PKY3 A0A310SJF4 B0W6N7 A0A0L7RJM9 V9IMB5 A0A067R1R4 A0A1Q3FA58 A0A182XX61 A0A0A9WSN1 A0A2A3EDH8 A0A146LYB2 E2B9F2 A0A088ADG8 A0A0C9RET2 A0A2J7PN84 E0VY83 D6WAL1 A0A1Y1MGZ3 A0A1E1XD65 A0A293L690 A0A1Z5LHR7 N6U1H1 A0A091S798 A0A0A6YVV7 A0A093KLA4 A0A3P4RZD5 A0A091QVD0 A0A1V4JXH3 A0A2D4IJS0 J3S5F7 A0A091IPM1 A0A087QVH0 A0A091G2N2
A0A2P8YKN3 K7J750 A0A1B6CBX0 A0A1B0D6S5 A0A182FSV7 A0A182LAH1 A0A3Q0J4I9 A0A182W135 A0A084W778 F5HJD2 A0A182RRT5 A0A1I8JVR1 A0A182M5P2 A0A182JFK9 A0A182SVX4 A0A1B6FDX3 A0A182I8L7 A0A0M8ZZF6 A0A0P6K128 A0A182K6A7 A0A154P1Z2 A0A182H264 A0A232FG18 T1JKE0 A0A240PKY3 A0A310SJF4 B0W6N7 A0A0L7RJM9 V9IMB5 A0A067R1R4 A0A1Q3FA58 A0A182XX61 A0A0A9WSN1 A0A2A3EDH8 A0A146LYB2 E2B9F2 A0A088ADG8 A0A0C9RET2 A0A2J7PN84 E0VY83 D6WAL1 A0A1Y1MGZ3 A0A1E1XD65 A0A293L690 A0A1Z5LHR7 N6U1H1 A0A091S798 A0A0A6YVV7 A0A093KLA4 A0A3P4RZD5 A0A091QVD0 A0A1V4JXH3 A0A2D4IJS0 J3S5F7 A0A091IPM1 A0A087QVH0 A0A091G2N2
Pubmed
EMBL
BABH01036433
BABH01036434
BABH01036435
KQ461108
KPJ08972.1
KQ459249
+ More
KPJ02217.1 AGBW02009261 OWR51229.1 GANO01001754 JAB58117.1 ADMH02002125 ETN58667.1 PYGN01000528 PSN44774.1 AAZX01008222 GEDC01026357 JAS10941.1 AJVK01026234 ATLV01021173 KE525314 KFB46072.1 AAAB01008844 EGK96393.1 AXCM01001964 GECZ01021343 JAS48426.1 APCN01003086 KQ435812 KOX72753.1 GDUN01000261 JAN95658.1 KQ434803 KZC05946.1 JXUM01104760 KQ564912 KXJ71580.1 NNAY01000297 OXU29379.1 AFFK01019697 KQ760549 OAD59875.1 DS231849 EDS36868.1 KQ414581 KOC71003.1 JR053449 AEY61932.1 KK852811 KDR15937.1 GFDL01010628 JAV24417.1 GBHO01033174 GBHO01033173 GDHC01008730 JAG10430.1 JAG10431.1 JAQ09899.1 KZ288271 PBC29833.1 GDHC01007003 JAQ11626.1 GL446502 EFN87676.1 GBYB01005496 JAG75263.1 NEVH01023957 PNF17804.1 DS235845 EEB18339.1 KQ971312 EEZ97982.1 GEZM01031084 GEZM01031083 JAV85072.1 GFAC01001996 JAT97192.1 GFWV01002787 MAA27517.1 GFJQ02000055 JAW06915.1 APGK01053202 KB741222 KB631665 ENN72402.1 ERL85146.1 KK944678 KFQ53406.1 AC084287 AC113597 KK556540 KFV98465.1 CYRY02045046 VCX40307.1 KK682873 KFQ13344.1 LSYS01005497 OPJ76823.1 IACK01101873 LAA84354.1 JU176434 AFJ51957.1 KL218035 KFP01611.1 KL225950 KFM05224.1 KL447642 KFO75606.1
KPJ02217.1 AGBW02009261 OWR51229.1 GANO01001754 JAB58117.1 ADMH02002125 ETN58667.1 PYGN01000528 PSN44774.1 AAZX01008222 GEDC01026357 JAS10941.1 AJVK01026234 ATLV01021173 KE525314 KFB46072.1 AAAB01008844 EGK96393.1 AXCM01001964 GECZ01021343 JAS48426.1 APCN01003086 KQ435812 KOX72753.1 GDUN01000261 JAN95658.1 KQ434803 KZC05946.1 JXUM01104760 KQ564912 KXJ71580.1 NNAY01000297 OXU29379.1 AFFK01019697 KQ760549 OAD59875.1 DS231849 EDS36868.1 KQ414581 KOC71003.1 JR053449 AEY61932.1 KK852811 KDR15937.1 GFDL01010628 JAV24417.1 GBHO01033174 GBHO01033173 GDHC01008730 JAG10430.1 JAG10431.1 JAQ09899.1 KZ288271 PBC29833.1 GDHC01007003 JAQ11626.1 GL446502 EFN87676.1 GBYB01005496 JAG75263.1 NEVH01023957 PNF17804.1 DS235845 EEB18339.1 KQ971312 EEZ97982.1 GEZM01031084 GEZM01031083 JAV85072.1 GFAC01001996 JAT97192.1 GFWV01002787 MAA27517.1 GFJQ02000055 JAW06915.1 APGK01053202 KB741222 KB631665 ENN72402.1 ERL85146.1 KK944678 KFQ53406.1 AC084287 AC113597 KK556540 KFV98465.1 CYRY02045046 VCX40307.1 KK682873 KFQ13344.1 LSYS01005497 OPJ76823.1 IACK01101873 LAA84354.1 JU176434 AFJ51957.1 KL218035 KFP01611.1 KL225950 KFM05224.1 KL447642 KFO75606.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000000673
UP000245037
+ More
UP000002358 UP000092462 UP000069272 UP000075882 UP000079169 UP000075920 UP000030765 UP000007062 UP000075900 UP000076407 UP000075883 UP000075880 UP000075901 UP000075840 UP000053105 UP000075881 UP000076502 UP000069940 UP000249989 UP000215335 UP000075885 UP000002320 UP000053825 UP000027135 UP000076408 UP000242457 UP000008237 UP000005203 UP000235965 UP000009046 UP000007266 UP000019118 UP000030742 UP000000589 UP000190648 UP000054308 UP000053286 UP000053760
UP000002358 UP000092462 UP000069272 UP000075882 UP000079169 UP000075920 UP000030765 UP000007062 UP000075900 UP000076407 UP000075883 UP000075880 UP000075901 UP000075840 UP000053105 UP000075881 UP000076502 UP000069940 UP000249989 UP000215335 UP000075885 UP000002320 UP000053825 UP000027135 UP000076408 UP000242457 UP000008237 UP000005203 UP000235965 UP000009046 UP000007266 UP000019118 UP000030742 UP000000589 UP000190648 UP000054308 UP000053286 UP000053760
PRIDE
Interpro
IPR036322
WD40_repeat_dom_sf
+ More
IPR001841 Znf_RING
IPR001680 WD40_repeat
IPR017986 WD40_repeat_dom
IPR013083 Znf_RING/FYVE/PHD
IPR017907 Znf_RING_CS
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR020472 G-protein_beta_WD-40_rep
IPR019775 WD40_repeat_CS
IPR003613 Ubox_domain
IPR000819 Peptidase_M17_C
IPR023042 Peptidase_M17_leu_NH2_pept
IPR011356 Leucine_aapep/pepB
IPR008283 Peptidase_M17_N
IPR001841 Znf_RING
IPR001680 WD40_repeat
IPR017986 WD40_repeat_dom
IPR013083 Znf_RING/FYVE/PHD
IPR017907 Znf_RING_CS
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR020472 G-protein_beta_WD-40_rep
IPR019775 WD40_repeat_CS
IPR003613 Ubox_domain
IPR000819 Peptidase_M17_C
IPR023042 Peptidase_M17_leu_NH2_pept
IPR011356 Leucine_aapep/pepB
IPR008283 Peptidase_M17_N
SUPFAM
SSF50978
SSF50978
Gene 3D
ProteinModelPortal
H9JJB9
A0A194QU24
A0A194Q9M7
A0A212FBW2
U5EVR0
W5J2X7
+ More
A0A2P8YKN3 K7J750 A0A1B6CBX0 A0A1B0D6S5 A0A182FSV7 A0A182LAH1 A0A3Q0J4I9 A0A182W135 A0A084W778 F5HJD2 A0A182RRT5 A0A1I8JVR1 A0A182M5P2 A0A182JFK9 A0A182SVX4 A0A1B6FDX3 A0A182I8L7 A0A0M8ZZF6 A0A0P6K128 A0A182K6A7 A0A154P1Z2 A0A182H264 A0A232FG18 T1JKE0 A0A240PKY3 A0A310SJF4 B0W6N7 A0A0L7RJM9 V9IMB5 A0A067R1R4 A0A1Q3FA58 A0A182XX61 A0A0A9WSN1 A0A2A3EDH8 A0A146LYB2 E2B9F2 A0A088ADG8 A0A0C9RET2 A0A2J7PN84 E0VY83 D6WAL1 A0A1Y1MGZ3 A0A1E1XD65 A0A293L690 A0A1Z5LHR7 N6U1H1 A0A091S798 A0A0A6YVV7 A0A093KLA4 A0A3P4RZD5 A0A091QVD0 A0A1V4JXH3 A0A2D4IJS0 J3S5F7 A0A091IPM1 A0A087QVH0 A0A091G2N2
A0A2P8YKN3 K7J750 A0A1B6CBX0 A0A1B0D6S5 A0A182FSV7 A0A182LAH1 A0A3Q0J4I9 A0A182W135 A0A084W778 F5HJD2 A0A182RRT5 A0A1I8JVR1 A0A182M5P2 A0A182JFK9 A0A182SVX4 A0A1B6FDX3 A0A182I8L7 A0A0M8ZZF6 A0A0P6K128 A0A182K6A7 A0A154P1Z2 A0A182H264 A0A232FG18 T1JKE0 A0A240PKY3 A0A310SJF4 B0W6N7 A0A0L7RJM9 V9IMB5 A0A067R1R4 A0A1Q3FA58 A0A182XX61 A0A0A9WSN1 A0A2A3EDH8 A0A146LYB2 E2B9F2 A0A088ADG8 A0A0C9RET2 A0A2J7PN84 E0VY83 D6WAL1 A0A1Y1MGZ3 A0A1E1XD65 A0A293L690 A0A1Z5LHR7 N6U1H1 A0A091S798 A0A0A6YVV7 A0A093KLA4 A0A3P4RZD5 A0A091QVD0 A0A1V4JXH3 A0A2D4IJS0 J3S5F7 A0A091IPM1 A0A087QVH0 A0A091G2N2
PDB
4R8P
E-value=4.0299e-05,
Score=110
Ontologies
GO
Topology
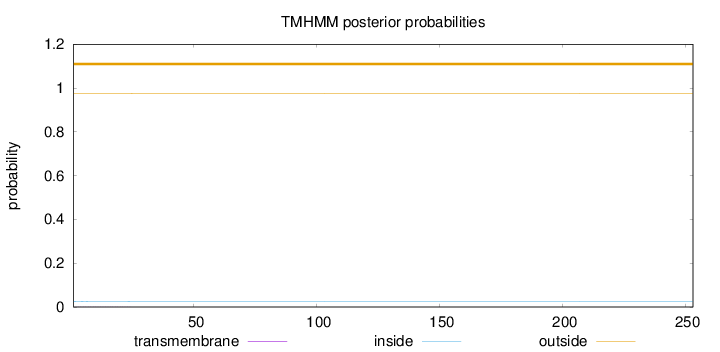
Length:
253
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01833
Exp number, first 60 AAs:
0.01815
Total prob of N-in:
0.02491
outside
1 - 253
Population Genetic Test Statistics
Pi
283.179072
Theta
193.421562
Tajima's D
1.323911
CLR
0
CSRT
0.746162691865407
Interpretation
Uncertain
