Pre Gene Modal
BGIBMGA009578
Annotation
PREDICTED:_putative_pre-mRNA-splicing_factor_ATP-dependent_RNA_helicase_PRP1_[Papilio_polytes]
Location in the cell
Cytoplasmic Reliability : 1.22 Mitochondrial Reliability : 1.326 Nuclear Reliability : 1.636
Sequence
CDS
ATGTCCAAACGTCGTATTGAAGTAATTGATCCGTTTATCAAAAAGAAAAGGGAAGAGAAGGCTGCGGCTGCTGCGAAATCTGGAATCACCGGTGAACCTTCAGAAGCAACTACCAACACGACTACCACCATGGCTACCAATACACCTGGACTAAATAAATATACCGGTCTTCCGCATTCACAGCGTTACCATGAGCTATTACGAAAACGTTTAGGACTCCCTGTCTGGGAATACAAAAATGATTTCATGAGATTACTCAACACTCATCAATGTATAGTGCTCGTAGGAGAAACAGGGTCAGGGAAGACTACCCAGATCCCCCAGTGGTCTGTAGAATTTGCTGCAGTAAGTGGCCTAGGCAAGGCTAAAGGAGTGGCCTGTACTCAACCAAGACGAGTGGCTGCTATGTCTGTTGCTCAGAGGGTAGCCGAAGAAATGGATGTGGCTTTGGGTCAGGAAGTTGGTTACAGCATTCGTTTTGAAGACTGTTCAGGGCCACAAACACTTCTAAAATACATGACAGACGGTATGTTGCTAAGAGAGGCTATGTCTGACCCAATGCTAGAACAATATAGAGTTATACTCCTCGATGAAGCCCACGAAAGGACACTTGCAACAGATATCCTGATGGGTGTTCTCAAAGAAGTCATAAAACAGAGGTCCGATTTGAAACTCGTTATTATGTCAGCCACATTAGATGCTGGAAAGTTCCAGCATTATTTTGACAATGCTCCATTAATGAACATTCCTGGTCGGACTCATCCGGTAGAAATATTTTACACTCCACAACCTGAACGAGATTACCTAGAAGCTGCGATAAGGACTGTAATACAAATCCATATGTGTGAAGAGATCTCTGGTGATATACTACTATTTTTAACTGGACAAGAGGAGATTGAGGATGCTTGTAAGAGAATTAAAAGAGAAATAGACAATCTGGGAACTGATGCAGGAGAGCTAAAGTGCATACCGTTATATTCAACATTGCCGCCAAATTTACAGCAACGGATCTTTGAGCCGGCACCTCCAAATCGACCAAACGGAGCCATCGGCAGGAAGGTGGTAGTGTCCACAAACATCGCTGAAACATCTCTGACTATTGATGGTGTTGTTTTTGTCATTGACCCAGGTTTTGCGAAACAAAAAGTTTATAATCCCCGTATAAGGGTGGAATCGTTATTGGTTTCTCCGATAAGTAAAGCATCAGCCCAACAGAGAGCGGGGAGGGCCGGCAGAACTAGACCTGGGAAGTGCTTCCGGTTATATACTGAGAAAGCATACAAGGATGAAATGCAGGACAATACTTATCCTGAGATTTTGAGATCAAACTTAGGATCTGTAGTTTTGCAGTTGAAGAAGTTGGGCATCGATGACCTGGTGCATTTCGATTTCATGGATCCACCGGCTCCGGAGACACTCATGCGGGCGCTCGAGCTGCTCAACTATCTGGCTGCGCTGGATGATGACGGTAACCTCACTGACCTGGGTGCTGTGATGGCGGAGTTCCCCCTCGACCCACAGCTGGCCAAGATGCTGATAGCCAGCTGCAATCACAACTGTTCCAACGAGATCCTCTCCATCACTGCCATGCTGTCAGTGCCGCAGTGCTTCGTGAGACCAAACGAGGCGCGGAAGGCTGCCGACGAGGCTAAAATGCGTTTTGCCCACATCGACGGTGACCATCTTACGCTACTGAACGTATACCACGCCTTCAAACAGAACATGGAGGATCCCCACTGGTGCTATGATAACTTCATTAATTACAGATCGCTAAAGTCGGGCGACAACGTCAGACAGCAACTTAGCAGGATTATGGACAGATTTAATTTGAAAAGAACAAGCACAGAATTCACAAGCAAAGATTATTACATCAACATACGGAAGGCACTTGTAAATGGATTTTTCATGCAGGTGGCGCACCTCGAGAGGACGGGCAGCTACCTGACGGTGAAGGACAACCAGGTGGTACAGCTGCACCCTTCCACGTGCCTGGACCACAAGCCCGACTGGGTCATATACAACGAGTTCGTGCTCACCACCAAGAACTACATTAGGACTGTCACTGATATCAAGCCGGAATGGCTGCTGAAGATCGCTCCCCAGTACTACGAGCTGGGCAACTTCCCGCAGTGCGAGGCGCGGAGACAGCTGGAGCTGCTGCAGGCCAGACTCGACTCCAAGCACTACCAGGAGGGCTTCTAG
Protein
MSKRRIEVIDPFIKKKREEKAAAAAKSGITGEPSEATTNTTTTMATNTPGLNKYTGLPHSQRYHELLRKRLGLPVWEYKNDFMRLLNTHQCIVLVGETGSGKTTQIPQWSVEFAAVSGLGKAKGVACTQPRRVAAMSVAQRVAEEMDVALGQEVGYSIRFEDCSGPQTLLKYMTDGMLLREAMSDPMLEQYRVILLDEAHERTLATDILMGVLKEVIKQRSDLKLVIMSATLDAGKFQHYFDNAPLMNIPGRTHPVEIFYTPQPERDYLEAAIRTVIQIHMCEEISGDILLFLTGQEEIEDACKRIKREIDNLGTDAGELKCIPLYSTLPPNLQQRIFEPAPPNRPNGAIGRKVVVSTNIAETSLTIDGVVFVIDPGFAKQKVYNPRIRVESLLVSPISKASAQQRAGRAGRTRPGKCFRLYTEKAYKDEMQDNTYPEILRSNLGSVVLQLKKLGIDDLVHFDFMDPPAPETLMRALELLNYLAALDDDGNLTDLGAVMAEFPLDPQLAKMLIASCNHNCSNEILSITAMLSVPQCFVRPNEARKAADEAKMRFAHIDGDHLTLLNVYHAFKQNMEDPHWCYDNFINYRSLKSGDNVRQQLSRIMDRFNLKRTSTEFTSKDYYINIRKALVNGFFMQVAHLERTGSYLTVKDNQVVQLHPSTCLDHKPDWVIYNEFVLTTKNYIRTVTDIKPEWLLKIAPQYYELGNFPQCEARRQLELLQARLDSKHYQEGF
Summary
Uniprot
H9JJ77
A0A194Q9Y3
S4NP01
A0A212FBX8
A0A0M9A9Y5
D6WA22
+ More
A0A1B6BZJ9 A0A088AGJ5 A0A067RNH9 A0A2J7QIQ9 N6UB91 A0A0N7Z9J6 K7IN36 A0A1B6F6H6 A0A0V0G8Z3 A0A023F5S5 A0A0C9RZM8 A0A158ND22 A0A154PFA4 A0A195B8Y2 A0A3L8DK02 A0A224XFJ3 A0A2A3EHY7 F4X0J9 A0A1I8MWF9 T1PAQ4 A0A0L0CT66 A0A0L7QVG8 A0A0A1XIY4 A0A1B0CGH3 A0A1L8DHD6 W8CAQ1 E0VIH1 E9IJS9 A0A1B0B1Z6 A0A026WH48 A0A151IDW4 E2AV08 A0A336LPD2 A0A195DUP8 A0A310S6W8 A0A1B0G6Z8 B4P1N2 A0A151XGJ3 A0A195F8M1 B3NA05 A0A3B0JD34 Q292S7 A0A0R3NL09 A0A034W103 B4GCN9 B4MR77 B4J6N1 A0A0M4E4B2 A0A1W4VE77 Q7K3M5 B4LM98 A0A0J9R727 B4HQT3 A0A182QBB9 E2C6Y1 U5ESQ2 A0A182YB18 A0A182N2A9 A0A2M4A757 A0A2M3Z2W7 A0A182FKL6 B4KNE3 A0A2M4BFR0 W5J7W3 A0A182RW62 B3MI69 A0A182VJV5 A0A182KL26 A0A182SFZ5 A0A1Y1LLP2 A0A182X5N1 A0A182JF92 A0A182TXC5 A0A182HWB7 A0A182NZM9 Q7QHE1 A0A182K266 T1IZ76 A0A232FAK2 J9K4C8 A0A2S2QEF0 Q17CT5 A0A084VHS4 A0A023EVY8 A0A2H8U1A7 A0A1Q3F2P6 B0WL58 A0A182MHD7 A0A023GLS7 A0A023FL45 A0A1E1XHM8 A0A1I8P3Q3 A0A1E1XTY7 A0A131YQK7
A0A1B6BZJ9 A0A088AGJ5 A0A067RNH9 A0A2J7QIQ9 N6UB91 A0A0N7Z9J6 K7IN36 A0A1B6F6H6 A0A0V0G8Z3 A0A023F5S5 A0A0C9RZM8 A0A158ND22 A0A154PFA4 A0A195B8Y2 A0A3L8DK02 A0A224XFJ3 A0A2A3EHY7 F4X0J9 A0A1I8MWF9 T1PAQ4 A0A0L0CT66 A0A0L7QVG8 A0A0A1XIY4 A0A1B0CGH3 A0A1L8DHD6 W8CAQ1 E0VIH1 E9IJS9 A0A1B0B1Z6 A0A026WH48 A0A151IDW4 E2AV08 A0A336LPD2 A0A195DUP8 A0A310S6W8 A0A1B0G6Z8 B4P1N2 A0A151XGJ3 A0A195F8M1 B3NA05 A0A3B0JD34 Q292S7 A0A0R3NL09 A0A034W103 B4GCN9 B4MR77 B4J6N1 A0A0M4E4B2 A0A1W4VE77 Q7K3M5 B4LM98 A0A0J9R727 B4HQT3 A0A182QBB9 E2C6Y1 U5ESQ2 A0A182YB18 A0A182N2A9 A0A2M4A757 A0A2M3Z2W7 A0A182FKL6 B4KNE3 A0A2M4BFR0 W5J7W3 A0A182RW62 B3MI69 A0A182VJV5 A0A182KL26 A0A182SFZ5 A0A1Y1LLP2 A0A182X5N1 A0A182JF92 A0A182TXC5 A0A182HWB7 A0A182NZM9 Q7QHE1 A0A182K266 T1IZ76 A0A232FAK2 J9K4C8 A0A2S2QEF0 Q17CT5 A0A084VHS4 A0A023EVY8 A0A2H8U1A7 A0A1Q3F2P6 B0WL58 A0A182MHD7 A0A023GLS7 A0A023FL45 A0A1E1XHM8 A0A1I8P3Q3 A0A1E1XTY7 A0A131YQK7
Pubmed
19121390
26354079
23622113
22118469
18362917
19820115
+ More
24845553 23537049 27129103 20075255 25474469 21347285 30249741 21719571 25315136 26108605 25830018 24495485 20566863 21282665 24508170 20798317 17994087 17550304 15632085 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25244985 20920257 23761445 20966253 28004739 12364791 28648823 17510324 24438588 24945155 26483478 28503490 29209593 26830274
24845553 23537049 27129103 20075255 25474469 21347285 30249741 21719571 25315136 26108605 25830018 24495485 20566863 21282665 24508170 20798317 17994087 17550304 15632085 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25244985 20920257 23761445 20966253 28004739 12364791 28648823 17510324 24438588 24945155 26483478 28503490 29209593 26830274
EMBL
BABH01036417
KQ459249
KPJ02229.1
GAIX01012059
JAA80501.1
AGBW02009261
+ More
OWR51233.1 KQ435706 KOX80227.1 KQ971312 EEZ98074.1 GEDC01030607 JAS06691.1 KK852563 KDR21284.1 NEVH01013579 PNF28459.1 APGK01030415 KB740771 KB632375 ENN78985.1 ERL93866.1 GDKW01000363 JAI56232.1 GECZ01024078 JAS45691.1 GECL01001924 JAP04200.1 GBBI01002185 JAC16527.1 GBYB01013586 JAG83353.1 ADTU01012186 KQ434892 KZC10529.1 KQ976558 KYM80654.1 QOIP01000007 RLU20503.1 GFTR01007858 JAW08568.1 KZ288235 PBC31413.1 GL888498 EGI59991.1 KA645784 AFP60413.1 JRES01000049 KNC34609.1 KQ414726 KOC62632.1 GBXI01003794 JAD10498.1 AJWK01011108 GFDF01008320 JAV05764.1 GAMC01005541 JAC01015.1 DS235199 EEB13177.1 GL763883 EFZ19125.1 JXJN01007391 KK107213 EZA55278.1 KQ977906 KYM98871.1 GL442974 EFN62726.1 UFQS01000094 UFQT01000094 SSW99275.1 SSX19655.1 KQ980341 KYN16477.1 KQ777390 OAD52096.1 CCAG010003750 CM000157 EDW89168.1 KQ982173 KYQ59418.1 KQ981744 KYN36399.1 CH954177 EDV59701.1 OUUW01000001 SPP73130.1 CM000071 EAL24784.1 KRT01556.1 GAKP01009726 JAC49226.1 CH479181 EDW31487.1 CH963849 EDW74616.1 CH916367 EDW00934.1 CP012524 ALC40841.1 AE013599 AY058484 AAF59269.1 AAL13713.1 AGB93299.1 CH940648 EDW60976.1 CM002911 KMY91952.1 CH480816 EDW46743.1 AXCN02001729 GL453236 EFN76299.1 GANO01003135 JAB56736.1 GGFK01003315 MBW36636.1 GGFM01002047 MBW22798.1 CH933808 EDW09996.1 GGFJ01002517 MBW51658.1 ADMH02001908 ETN60552.1 CH902619 EDV35914.1 GEZM01054093 JAV73851.1 APCN01001484 AAAB01008816 EAA05149.4 AFFK01020413 NNAY01000599 OXU27468.1 ABLF02019760 ABLF02053299 GGMS01006914 MBY76117.1 CH477304 EAT44187.1 ATLV01013224 KE524847 KFB37518.1 JXUM01033610 JXUM01126431 GAPW01000337 KQ567067 KQ560995 JAC13261.1 KXJ69639.1 KXJ80091.1 GFXV01008026 MBW19831.1 GFDL01013277 JAV21768.1 DS231980 EDS30231.1 AXCM01007440 GBBM01000561 JAC34857.1 GBBK01002130 JAC22352.1 GFAC01000485 JAT98703.1 GFAA01001013 JAU02422.1 GEDV01007018 JAP81539.1
OWR51233.1 KQ435706 KOX80227.1 KQ971312 EEZ98074.1 GEDC01030607 JAS06691.1 KK852563 KDR21284.1 NEVH01013579 PNF28459.1 APGK01030415 KB740771 KB632375 ENN78985.1 ERL93866.1 GDKW01000363 JAI56232.1 GECZ01024078 JAS45691.1 GECL01001924 JAP04200.1 GBBI01002185 JAC16527.1 GBYB01013586 JAG83353.1 ADTU01012186 KQ434892 KZC10529.1 KQ976558 KYM80654.1 QOIP01000007 RLU20503.1 GFTR01007858 JAW08568.1 KZ288235 PBC31413.1 GL888498 EGI59991.1 KA645784 AFP60413.1 JRES01000049 KNC34609.1 KQ414726 KOC62632.1 GBXI01003794 JAD10498.1 AJWK01011108 GFDF01008320 JAV05764.1 GAMC01005541 JAC01015.1 DS235199 EEB13177.1 GL763883 EFZ19125.1 JXJN01007391 KK107213 EZA55278.1 KQ977906 KYM98871.1 GL442974 EFN62726.1 UFQS01000094 UFQT01000094 SSW99275.1 SSX19655.1 KQ980341 KYN16477.1 KQ777390 OAD52096.1 CCAG010003750 CM000157 EDW89168.1 KQ982173 KYQ59418.1 KQ981744 KYN36399.1 CH954177 EDV59701.1 OUUW01000001 SPP73130.1 CM000071 EAL24784.1 KRT01556.1 GAKP01009726 JAC49226.1 CH479181 EDW31487.1 CH963849 EDW74616.1 CH916367 EDW00934.1 CP012524 ALC40841.1 AE013599 AY058484 AAF59269.1 AAL13713.1 AGB93299.1 CH940648 EDW60976.1 CM002911 KMY91952.1 CH480816 EDW46743.1 AXCN02001729 GL453236 EFN76299.1 GANO01003135 JAB56736.1 GGFK01003315 MBW36636.1 GGFM01002047 MBW22798.1 CH933808 EDW09996.1 GGFJ01002517 MBW51658.1 ADMH02001908 ETN60552.1 CH902619 EDV35914.1 GEZM01054093 JAV73851.1 APCN01001484 AAAB01008816 EAA05149.4 AFFK01020413 NNAY01000599 OXU27468.1 ABLF02019760 ABLF02053299 GGMS01006914 MBY76117.1 CH477304 EAT44187.1 ATLV01013224 KE524847 KFB37518.1 JXUM01033610 JXUM01126431 GAPW01000337 KQ567067 KQ560995 JAC13261.1 KXJ69639.1 KXJ80091.1 GFXV01008026 MBW19831.1 GFDL01013277 JAV21768.1 DS231980 EDS30231.1 AXCM01007440 GBBM01000561 JAC34857.1 GBBK01002130 JAC22352.1 GFAC01000485 JAT98703.1 GFAA01001013 JAU02422.1 GEDV01007018 JAP81539.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000053105
UP000007266
UP000005203
+ More
UP000027135 UP000235965 UP000019118 UP000030742 UP000002358 UP000005205 UP000076502 UP000078540 UP000279307 UP000242457 UP000007755 UP000095301 UP000037069 UP000053825 UP000092461 UP000009046 UP000092460 UP000053097 UP000078542 UP000000311 UP000078492 UP000092444 UP000002282 UP000075809 UP000078541 UP000008711 UP000268350 UP000001819 UP000008744 UP000007798 UP000001070 UP000092553 UP000192221 UP000000803 UP000008792 UP000001292 UP000075886 UP000008237 UP000076408 UP000075884 UP000069272 UP000009192 UP000000673 UP000075900 UP000007801 UP000075903 UP000075882 UP000075901 UP000076407 UP000075880 UP000075902 UP000075840 UP000075885 UP000007062 UP000075881 UP000215335 UP000007819 UP000008820 UP000030765 UP000069940 UP000249989 UP000002320 UP000075883 UP000095300
UP000027135 UP000235965 UP000019118 UP000030742 UP000002358 UP000005205 UP000076502 UP000078540 UP000279307 UP000242457 UP000007755 UP000095301 UP000037069 UP000053825 UP000092461 UP000009046 UP000092460 UP000053097 UP000078542 UP000000311 UP000078492 UP000092444 UP000002282 UP000075809 UP000078541 UP000008711 UP000268350 UP000001819 UP000008744 UP000007798 UP000001070 UP000092553 UP000192221 UP000000803 UP000008792 UP000001292 UP000075886 UP000008237 UP000076408 UP000075884 UP000069272 UP000009192 UP000000673 UP000075900 UP000007801 UP000075903 UP000075882 UP000075901 UP000076407 UP000075880 UP000075902 UP000075840 UP000075885 UP000007062 UP000075881 UP000215335 UP000007819 UP000008820 UP000030765 UP000069940 UP000249989 UP000002320 UP000075883 UP000095300
Interpro
IPR001650
Helicase_C
+ More
IPR014001 Helicase_ATP-bd
IPR011709 DUF1605
IPR007502 Helicase-assoc_dom
IPR027417 P-loop_NTPase
IPR011545 DEAD/DEAH_box_helicase_dom
IPR001609 Myosin_head_motor_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR019775 WD40_repeat_CS
IPR036322 WD40_repeat_dom_sf
IPR014014 RNA_helicase_DEAD_Q_motif
IPR002464 DNA/RNA_helicase_DEAH_CS
IPR014001 Helicase_ATP-bd
IPR011709 DUF1605
IPR007502 Helicase-assoc_dom
IPR027417 P-loop_NTPase
IPR011545 DEAD/DEAH_box_helicase_dom
IPR001609 Myosin_head_motor_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR019775 WD40_repeat_CS
IPR036322 WD40_repeat_dom_sf
IPR014014 RNA_helicase_DEAD_Q_motif
IPR002464 DNA/RNA_helicase_DEAH_CS
Gene 3D
CDD
ProteinModelPortal
H9JJ77
A0A194Q9Y3
S4NP01
A0A212FBX8
A0A0M9A9Y5
D6WA22
+ More
A0A1B6BZJ9 A0A088AGJ5 A0A067RNH9 A0A2J7QIQ9 N6UB91 A0A0N7Z9J6 K7IN36 A0A1B6F6H6 A0A0V0G8Z3 A0A023F5S5 A0A0C9RZM8 A0A158ND22 A0A154PFA4 A0A195B8Y2 A0A3L8DK02 A0A224XFJ3 A0A2A3EHY7 F4X0J9 A0A1I8MWF9 T1PAQ4 A0A0L0CT66 A0A0L7QVG8 A0A0A1XIY4 A0A1B0CGH3 A0A1L8DHD6 W8CAQ1 E0VIH1 E9IJS9 A0A1B0B1Z6 A0A026WH48 A0A151IDW4 E2AV08 A0A336LPD2 A0A195DUP8 A0A310S6W8 A0A1B0G6Z8 B4P1N2 A0A151XGJ3 A0A195F8M1 B3NA05 A0A3B0JD34 Q292S7 A0A0R3NL09 A0A034W103 B4GCN9 B4MR77 B4J6N1 A0A0M4E4B2 A0A1W4VE77 Q7K3M5 B4LM98 A0A0J9R727 B4HQT3 A0A182QBB9 E2C6Y1 U5ESQ2 A0A182YB18 A0A182N2A9 A0A2M4A757 A0A2M3Z2W7 A0A182FKL6 B4KNE3 A0A2M4BFR0 W5J7W3 A0A182RW62 B3MI69 A0A182VJV5 A0A182KL26 A0A182SFZ5 A0A1Y1LLP2 A0A182X5N1 A0A182JF92 A0A182TXC5 A0A182HWB7 A0A182NZM9 Q7QHE1 A0A182K266 T1IZ76 A0A232FAK2 J9K4C8 A0A2S2QEF0 Q17CT5 A0A084VHS4 A0A023EVY8 A0A2H8U1A7 A0A1Q3F2P6 B0WL58 A0A182MHD7 A0A023GLS7 A0A023FL45 A0A1E1XHM8 A0A1I8P3Q3 A0A1E1XTY7 A0A131YQK7
A0A1B6BZJ9 A0A088AGJ5 A0A067RNH9 A0A2J7QIQ9 N6UB91 A0A0N7Z9J6 K7IN36 A0A1B6F6H6 A0A0V0G8Z3 A0A023F5S5 A0A0C9RZM8 A0A158ND22 A0A154PFA4 A0A195B8Y2 A0A3L8DK02 A0A224XFJ3 A0A2A3EHY7 F4X0J9 A0A1I8MWF9 T1PAQ4 A0A0L0CT66 A0A0L7QVG8 A0A0A1XIY4 A0A1B0CGH3 A0A1L8DHD6 W8CAQ1 E0VIH1 E9IJS9 A0A1B0B1Z6 A0A026WH48 A0A151IDW4 E2AV08 A0A336LPD2 A0A195DUP8 A0A310S6W8 A0A1B0G6Z8 B4P1N2 A0A151XGJ3 A0A195F8M1 B3NA05 A0A3B0JD34 Q292S7 A0A0R3NL09 A0A034W103 B4GCN9 B4MR77 B4J6N1 A0A0M4E4B2 A0A1W4VE77 Q7K3M5 B4LM98 A0A0J9R727 B4HQT3 A0A182QBB9 E2C6Y1 U5ESQ2 A0A182YB18 A0A182N2A9 A0A2M4A757 A0A2M3Z2W7 A0A182FKL6 B4KNE3 A0A2M4BFR0 W5J7W3 A0A182RW62 B3MI69 A0A182VJV5 A0A182KL26 A0A182SFZ5 A0A1Y1LLP2 A0A182X5N1 A0A182JF92 A0A182TXC5 A0A182HWB7 A0A182NZM9 Q7QHE1 A0A182K266 T1IZ76 A0A232FAK2 J9K4C8 A0A2S2QEF0 Q17CT5 A0A084VHS4 A0A023EVY8 A0A2H8U1A7 A0A1Q3F2P6 B0WL58 A0A182MHD7 A0A023GLS7 A0A023FL45 A0A1E1XHM8 A0A1I8P3Q3 A0A1E1XTY7 A0A131YQK7
PDB
6ID1
E-value=0,
Score=2979
Ontologies
GO
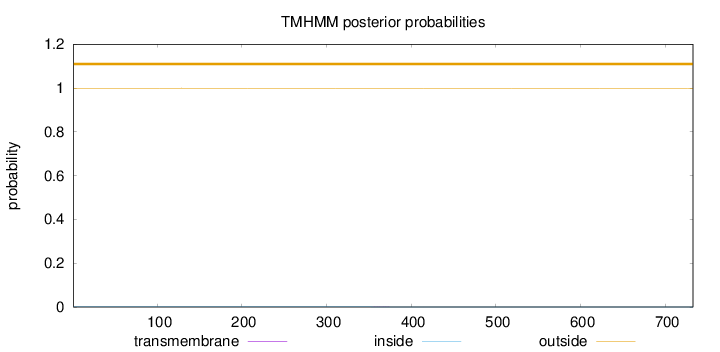
Topology
Length:
733
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00483999999999999
Exp number, first 60 AAs:
0.00016
Total prob of N-in:
0.00064
outside
1 - 733
Population Genetic Test Statistics
Pi
321.82207
Theta
186.86046
Tajima's D
2.082901
CLR
0.000019
CSRT
0.890705464726764
Interpretation
Uncertain
