Pre Gene Modal
BGIBMGA009618
Annotation
PREDICTED:_grpE_protein_homolog?_mitochondrial_[Amyelois_transitella]
Full name
GrpE protein homolog
+ More
GrpE protein homolog, mitochondrial
GrpE protein homolog, mitochondrial
Alternative Name
dRoe1
Location in the cell
Mitochondrial Reliability : 2.707
Sequence
CDS
ATGGCTCTAAAAGCTTATACATTTGGGAGAATAGCAAAATACTTGGCGGATACCACGCTGAGACCAAATCTAACGATCAAAACTTTCAGACCATCCGTTCGTGCGCTCAGTGCAGAGAAGCCAGCCGAGTCGGAGTCAAATGACAAGCTCCCGGCCACCATAGAGGAATGTCATAAACAAATTGAAAGTTTGACTAATGAAGTTAATTCTATTAAAGAACAATTCAATGACATTAATGACAAATACAAGAGAGCGTTAGCGGACGGCGAGAATGTACGGAAGAGGATGTTGAAACAAGTCGAAGACGCTAAGTCCTTTGCCATACAGAGCTTCTGCAAGGATCTGCTAGAGGTAGCAGACACATTGACGGCTGCAGCAGAGAGCATACCAGAGGGAGCGGGGGCGGAGGCCGAGGGCGGCGCCCTACGAGCCCTGCACGACGGTGTGCGACTCACGAGGGCTCAACTAGGACAGGTGTTCGCGCGGCACGGCCTGGTCCCGGTGTCTCCCCTCCGAGAGAAATTCGACCCGAATCTACATGAAGCATTGTTCCAGCAGGAAGTGGAGGGCGCCGAGTCAGGCACAGTGGTGGCCGTCAGCAAGGTCGGCTACAAACTGCACGAGAGATGCGTCAGACCAGCTTTGGTTGGGGTGGCGAAATAA
Protein
MALKAYTFGRIAKYLADTTLRPNLTIKTFRPSVRALSAEKPAESESNDKLPATIEECHKQIESLTNEVNSIKEQFNDINDKYKRALADGENVRKRMLKQVEDAKSFAIQSFCKDLLEVADTLTAAAESIPEGAGAEAEGGALRALHDGVRLTRAQLGQVFARHGLVPVSPLREKFDPNLHEALFQQEVEGAESGTVVAVSKVGYKLHERCVRPALVGVAK
Summary
Description
Essential component of the PAM complex, a complex required for the translocation of transit peptide-containing proteins from the inner membrane into the mitochondrial matrix in an ATP-dependent manner.
Essential component of the PAM complex, a complex required for the translocation of transit peptide-containing proteins from the inner membrane into the mitochondrial matrix in an ATP-dependent manner. Seems to control the nucleotide-dependent binding of mitochondrial HSP70 to substrate proteins (By similarity).
Essential component of the PAM complex, a complex required for the translocation of transit peptide-containing proteins from the inner membrane into the mitochondrial matrix in an ATP-dependent manner. Seems to control the nucleotide-dependent binding of mitochondrial HSP70 to substrate proteins (By similarity).
Subunit
Probable component of the PAM complex at least composed of a mitochondrial HSP70 protein, Roe1, TIM44, blp/TIM16 and TIM14.
Similarity
Belongs to the GrpE family.
Keywords
Chaperone
Complete proteome
Mitochondrion
Reference proteome
Transit peptide
Feature
chain GrpE protein homolog
Uniprot
H9JJB7
I4DMZ3
A0A0N1PK10
A0A2H1VMM4
A0A194QB61
A0A212FBX7
+ More
A0A2A4JSC2 A0A0K8SQV7 A0A0A9YND4 A0A1B6CDT7 U5EZP0 A0A146MC23 A0A146KYD7 R4V2H8 Q17CT4 E0VIG9 A0A164VPI4 A0A2M3Z902 A0A2M4BXP0 A0A023EMJ3 A0A182HCP9 A0A0N8CUT5 A0A0P5U7S6 A0A0P5THI2 A0A0P5X4Y6 A0A182FKL7 K7IN38 A0A0P5C365 A0A2M3Z942 A0A0L7QVR3 A0A0P5KGD4 A0A2M3Z915 A0A232F9E0 A0A2M4AM83 A0A2M4ALE0 A0A2M4AI43 A0A2M4API4 A0A2M4AI24 A0A2M4ANY9 A0A0P6HBW2 A0A2R7W824 W5JAW9 A0A1Q3F9Q8 A0A2M4BXM3 A0A1Y1N326 A0A2M4BXN8 A0A0P5BPD2 A0A088AGJ6 A0A0P6AL19 A0A310SCQ6 A0A067RL06 B0WL57 B4HQK8 B4KMJ8 A0A224XWW6 A0A0P4VMI1 T1HP54 A0A2A3EIW0 A0A0C9QRM5 B6IDM7 P48604 B4QEF7 V9IGZ1 B4P4F7 W8B6J8 B3NRJ1 A0A0V0G5J4 E9FXR3 A0A154PFD7 A0A034VZK2 A0A0K8TNL8 A0A0K8VKQ3 A0A182YB19 A0A0J7KRP3 A0A023F901 A0A026WE19 A0A3L8E276 B4MCN0 D6WBZ8 A0A195CXI8 A0A0N0U7N1 B4JVP2 A0A151JCI4 B3MHC4 A0A182N2A8 J3JWN2 A0A182J0D2 A0A1B0D6T2 A0A1W4VGQ8 A0A182X5N0 E2AKF8 B4MS96 A0A182K267 E9IZB2 A0A195AUE3 A0A158P0T5 Q28ZS1 Q7QHE2 A0A182HWB8
A0A2A4JSC2 A0A0K8SQV7 A0A0A9YND4 A0A1B6CDT7 U5EZP0 A0A146MC23 A0A146KYD7 R4V2H8 Q17CT4 E0VIG9 A0A164VPI4 A0A2M3Z902 A0A2M4BXP0 A0A023EMJ3 A0A182HCP9 A0A0N8CUT5 A0A0P5U7S6 A0A0P5THI2 A0A0P5X4Y6 A0A182FKL7 K7IN38 A0A0P5C365 A0A2M3Z942 A0A0L7QVR3 A0A0P5KGD4 A0A2M3Z915 A0A232F9E0 A0A2M4AM83 A0A2M4ALE0 A0A2M4AI43 A0A2M4API4 A0A2M4AI24 A0A2M4ANY9 A0A0P6HBW2 A0A2R7W824 W5JAW9 A0A1Q3F9Q8 A0A2M4BXM3 A0A1Y1N326 A0A2M4BXN8 A0A0P5BPD2 A0A088AGJ6 A0A0P6AL19 A0A310SCQ6 A0A067RL06 B0WL57 B4HQK8 B4KMJ8 A0A224XWW6 A0A0P4VMI1 T1HP54 A0A2A3EIW0 A0A0C9QRM5 B6IDM7 P48604 B4QEF7 V9IGZ1 B4P4F7 W8B6J8 B3NRJ1 A0A0V0G5J4 E9FXR3 A0A154PFD7 A0A034VZK2 A0A0K8TNL8 A0A0K8VKQ3 A0A182YB19 A0A0J7KRP3 A0A023F901 A0A026WE19 A0A3L8E276 B4MCN0 D6WBZ8 A0A195CXI8 A0A0N0U7N1 B4JVP2 A0A151JCI4 B3MHC4 A0A182N2A8 J3JWN2 A0A182J0D2 A0A1B0D6T2 A0A1W4VGQ8 A0A182X5N0 E2AKF8 B4MS96 A0A182K267 E9IZB2 A0A195AUE3 A0A158P0T5 Q28ZS1 Q7QHE2 A0A182HWB8
Pubmed
19121390
22651552
26354079
22118469
25401762
26823975
+ More
17510324 20566863 24945155 26483478 20075255 28648823 20920257 23761445 28004739 24845553 17994087 27129103 10731132 12537572 12537569 22936249 17550304 24495485 21292972 25348373 26369729 25244985 25474469 24508170 30249741 18362917 19820115 22516182 23537049 20798317 21282665 21347285 15632085 12364791
17510324 20566863 24945155 26483478 20075255 28648823 20920257 23761445 28004739 24845553 17994087 27129103 10731132 12537572 12537569 22936249 17550304 24495485 21292972 25348373 26369729 25244985 25474469 24508170 30249741 18362917 19820115 22516182 23537049 20798317 21282665 21347285 15632085 12364791
EMBL
BABH01036417
AK402661
BAM19283.1
LADJ01051845
KPJ21509.1
ODYU01003372
+ More
SOQ42046.1 KQ459249 KPJ02230.1 AGBW02009261 OWR51235.1 NWSH01000762 PCG74363.1 GBRD01010228 JAG55596.1 GBHO01012559 JAG31045.1 GEDC01025684 JAS11614.1 GANO01000431 JAB59440.1 GDHC01001640 JAQ16989.1 GDHC01018592 JAQ00037.1 KC741113 AGM32937.1 CH477304 EAT44188.1 DS235199 EEB13175.1 LRGB01001361 KZS12524.1 GGFM01004252 MBW25003.1 GGFJ01008688 MBW57829.1 GAPW01004054 JAC09544.1 JXUM01033610 KQ560995 KXJ80092.1 GDIP01096767 JAM06948.1 GDIP01116913 JAL86801.1 GDIP01126164 JAL77550.1 GDIP01077417 JAM26298.1 GDIP01180356 JAJ43046.1 GGFM01004293 MBW25044.1 KQ414726 KOC62631.1 GDIQ01184581 GDIQ01184580 JAK67144.1 GGFM01004276 MBW25027.1 NNAY01000599 OXU27471.1 GGFK01008569 MBW41890.1 GGFK01008282 MBW41603.1 GGFK01007123 MBW40444.1 GGFK01009346 MBW42667.1 GGFK01007103 MBW40424.1 GGFK01009178 MBW42499.1 GDIQ01021917 JAN72820.1 KK854300 PTY14355.1 ADMH02001908 ETN60553.1 GFDL01010796 JAV24249.1 GGFJ01008686 MBW57827.1 GEZM01013860 JAV92283.1 GGFJ01008689 MBW57830.1 GDIP01182104 JAJ41298.1 GDIP01027816 JAM75899.1 KQ777390 OAD52097.1 KK852563 KDR21285.1 DS231980 EDS30230.1 CH480816 EDW47743.1 CH933808 EDW10845.2 GFTR01003897 JAW12529.1 GDKW01000844 JAI55751.1 ACPB03001795 KZ288235 PBC31414.1 GBYB01003282 JAG73049.1 BT050467 ACJ13174.1 U34903 AE013599 AY094889 CM000362 CM002911 EDX06947.1 KMY93539.1 JR047325 AEY60378.1 CM000158 EDW90596.1 GAMC01009745 JAB96810.1 CH954179 EDV56143.1 GECL01002863 JAP03261.1 GL732526 EFX88242.1 KQ434892 KZC10527.1 GAKP01011098 JAC47854.1 GDAI01001616 JAI15987.1 GDHF01013174 JAI39140.1 LBMM01003943 KMQ92966.1 GBBI01001273 JAC17439.1 KK107260 EZA54163.1 QOIP01000001 RLU26622.1 CH940659 EDW71418.2 KQ971309 EEZ99113.1 KQ977141 KYN05380.1 KQ435706 KOX80228.1 CH916375 EDV98510.1 KQ979038 KYN23464.1 CH902619 EDV37924.1 BT127650 KB632375 AEE62612.1 ERL94032.1 AJVK01026253 GL440262 EFN66100.1 CH963850 EDW74985.2 GL767121 EFZ14048.1 KQ976738 KYM75677.1 ADTU01005782 ADTU01005783 CM000071 EAL25542.1 AAAB01008816 EAA05028.4 APCN01001484
SOQ42046.1 KQ459249 KPJ02230.1 AGBW02009261 OWR51235.1 NWSH01000762 PCG74363.1 GBRD01010228 JAG55596.1 GBHO01012559 JAG31045.1 GEDC01025684 JAS11614.1 GANO01000431 JAB59440.1 GDHC01001640 JAQ16989.1 GDHC01018592 JAQ00037.1 KC741113 AGM32937.1 CH477304 EAT44188.1 DS235199 EEB13175.1 LRGB01001361 KZS12524.1 GGFM01004252 MBW25003.1 GGFJ01008688 MBW57829.1 GAPW01004054 JAC09544.1 JXUM01033610 KQ560995 KXJ80092.1 GDIP01096767 JAM06948.1 GDIP01116913 JAL86801.1 GDIP01126164 JAL77550.1 GDIP01077417 JAM26298.1 GDIP01180356 JAJ43046.1 GGFM01004293 MBW25044.1 KQ414726 KOC62631.1 GDIQ01184581 GDIQ01184580 JAK67144.1 GGFM01004276 MBW25027.1 NNAY01000599 OXU27471.1 GGFK01008569 MBW41890.1 GGFK01008282 MBW41603.1 GGFK01007123 MBW40444.1 GGFK01009346 MBW42667.1 GGFK01007103 MBW40424.1 GGFK01009178 MBW42499.1 GDIQ01021917 JAN72820.1 KK854300 PTY14355.1 ADMH02001908 ETN60553.1 GFDL01010796 JAV24249.1 GGFJ01008686 MBW57827.1 GEZM01013860 JAV92283.1 GGFJ01008689 MBW57830.1 GDIP01182104 JAJ41298.1 GDIP01027816 JAM75899.1 KQ777390 OAD52097.1 KK852563 KDR21285.1 DS231980 EDS30230.1 CH480816 EDW47743.1 CH933808 EDW10845.2 GFTR01003897 JAW12529.1 GDKW01000844 JAI55751.1 ACPB03001795 KZ288235 PBC31414.1 GBYB01003282 JAG73049.1 BT050467 ACJ13174.1 U34903 AE013599 AY094889 CM000362 CM002911 EDX06947.1 KMY93539.1 JR047325 AEY60378.1 CM000158 EDW90596.1 GAMC01009745 JAB96810.1 CH954179 EDV56143.1 GECL01002863 JAP03261.1 GL732526 EFX88242.1 KQ434892 KZC10527.1 GAKP01011098 JAC47854.1 GDAI01001616 JAI15987.1 GDHF01013174 JAI39140.1 LBMM01003943 KMQ92966.1 GBBI01001273 JAC17439.1 KK107260 EZA54163.1 QOIP01000001 RLU26622.1 CH940659 EDW71418.2 KQ971309 EEZ99113.1 KQ977141 KYN05380.1 KQ435706 KOX80228.1 CH916375 EDV98510.1 KQ979038 KYN23464.1 CH902619 EDV37924.1 BT127650 KB632375 AEE62612.1 ERL94032.1 AJVK01026253 GL440262 EFN66100.1 CH963850 EDW74985.2 GL767121 EFZ14048.1 KQ976738 KYM75677.1 ADTU01005782 ADTU01005783 CM000071 EAL25542.1 AAAB01008816 EAA05028.4 APCN01001484
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000218220
UP000008820
+ More
UP000009046 UP000076858 UP000069940 UP000249989 UP000069272 UP000002358 UP000053825 UP000215335 UP000000673 UP000005203 UP000027135 UP000002320 UP000001292 UP000009192 UP000015103 UP000242457 UP000000803 UP000000304 UP000002282 UP000008711 UP000000305 UP000076502 UP000076408 UP000036403 UP000053097 UP000279307 UP000008792 UP000007266 UP000078542 UP000053105 UP000001070 UP000078492 UP000007801 UP000075884 UP000030742 UP000075880 UP000092462 UP000192221 UP000076407 UP000000311 UP000007798 UP000075881 UP000078540 UP000005205 UP000001819 UP000007062 UP000075840
UP000009046 UP000076858 UP000069940 UP000249989 UP000069272 UP000002358 UP000053825 UP000215335 UP000000673 UP000005203 UP000027135 UP000002320 UP000001292 UP000009192 UP000015103 UP000242457 UP000000803 UP000000304 UP000002282 UP000008711 UP000000305 UP000076502 UP000076408 UP000036403 UP000053097 UP000279307 UP000008792 UP000007266 UP000078542 UP000053105 UP000001070 UP000078492 UP000007801 UP000075884 UP000030742 UP000075880 UP000092462 UP000192221 UP000076407 UP000000311 UP000007798 UP000075881 UP000078540 UP000005205 UP000001819 UP000007062 UP000075840
Pfam
PF01025 GrpE
Gene 3D
CDD
ProteinModelPortal
H9JJB7
I4DMZ3
A0A0N1PK10
A0A2H1VMM4
A0A194QB61
A0A212FBX7
+ More
A0A2A4JSC2 A0A0K8SQV7 A0A0A9YND4 A0A1B6CDT7 U5EZP0 A0A146MC23 A0A146KYD7 R4V2H8 Q17CT4 E0VIG9 A0A164VPI4 A0A2M3Z902 A0A2M4BXP0 A0A023EMJ3 A0A182HCP9 A0A0N8CUT5 A0A0P5U7S6 A0A0P5THI2 A0A0P5X4Y6 A0A182FKL7 K7IN38 A0A0P5C365 A0A2M3Z942 A0A0L7QVR3 A0A0P5KGD4 A0A2M3Z915 A0A232F9E0 A0A2M4AM83 A0A2M4ALE0 A0A2M4AI43 A0A2M4API4 A0A2M4AI24 A0A2M4ANY9 A0A0P6HBW2 A0A2R7W824 W5JAW9 A0A1Q3F9Q8 A0A2M4BXM3 A0A1Y1N326 A0A2M4BXN8 A0A0P5BPD2 A0A088AGJ6 A0A0P6AL19 A0A310SCQ6 A0A067RL06 B0WL57 B4HQK8 B4KMJ8 A0A224XWW6 A0A0P4VMI1 T1HP54 A0A2A3EIW0 A0A0C9QRM5 B6IDM7 P48604 B4QEF7 V9IGZ1 B4P4F7 W8B6J8 B3NRJ1 A0A0V0G5J4 E9FXR3 A0A154PFD7 A0A034VZK2 A0A0K8TNL8 A0A0K8VKQ3 A0A182YB19 A0A0J7KRP3 A0A023F901 A0A026WE19 A0A3L8E276 B4MCN0 D6WBZ8 A0A195CXI8 A0A0N0U7N1 B4JVP2 A0A151JCI4 B3MHC4 A0A182N2A8 J3JWN2 A0A182J0D2 A0A1B0D6T2 A0A1W4VGQ8 A0A182X5N0 E2AKF8 B4MS96 A0A182K267 E9IZB2 A0A195AUE3 A0A158P0T5 Q28ZS1 Q7QHE2 A0A182HWB8
A0A2A4JSC2 A0A0K8SQV7 A0A0A9YND4 A0A1B6CDT7 U5EZP0 A0A146MC23 A0A146KYD7 R4V2H8 Q17CT4 E0VIG9 A0A164VPI4 A0A2M3Z902 A0A2M4BXP0 A0A023EMJ3 A0A182HCP9 A0A0N8CUT5 A0A0P5U7S6 A0A0P5THI2 A0A0P5X4Y6 A0A182FKL7 K7IN38 A0A0P5C365 A0A2M3Z942 A0A0L7QVR3 A0A0P5KGD4 A0A2M3Z915 A0A232F9E0 A0A2M4AM83 A0A2M4ALE0 A0A2M4AI43 A0A2M4API4 A0A2M4AI24 A0A2M4ANY9 A0A0P6HBW2 A0A2R7W824 W5JAW9 A0A1Q3F9Q8 A0A2M4BXM3 A0A1Y1N326 A0A2M4BXN8 A0A0P5BPD2 A0A088AGJ6 A0A0P6AL19 A0A310SCQ6 A0A067RL06 B0WL57 B4HQK8 B4KMJ8 A0A224XWW6 A0A0P4VMI1 T1HP54 A0A2A3EIW0 A0A0C9QRM5 B6IDM7 P48604 B4QEF7 V9IGZ1 B4P4F7 W8B6J8 B3NRJ1 A0A0V0G5J4 E9FXR3 A0A154PFD7 A0A034VZK2 A0A0K8TNL8 A0A0K8VKQ3 A0A182YB19 A0A0J7KRP3 A0A023F901 A0A026WE19 A0A3L8E276 B4MCN0 D6WBZ8 A0A195CXI8 A0A0N0U7N1 B4JVP2 A0A151JCI4 B3MHC4 A0A182N2A8 J3JWN2 A0A182J0D2 A0A1B0D6T2 A0A1W4VGQ8 A0A182X5N0 E2AKF8 B4MS96 A0A182K267 E9IZB2 A0A195AUE3 A0A158P0T5 Q28ZS1 Q7QHE2 A0A182HWB8
PDB
4ANI
E-value=4.64311e-13,
Score=177
Ontologies
GO
PANTHER
Topology
Subcellular location
Mitochondrion matrix
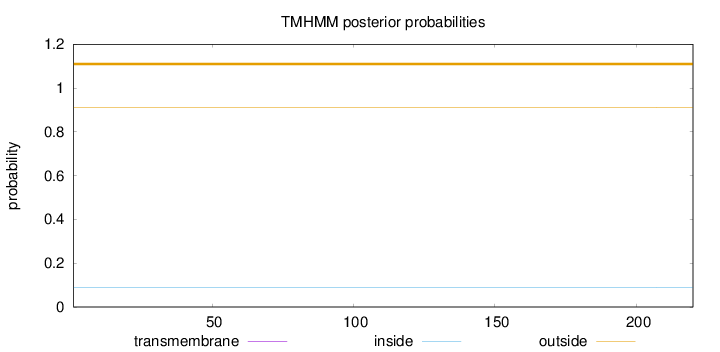
Length:
220
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00016
Exp number, first 60 AAs:
0.00016
Total prob of N-in:
0.08799
outside
1 - 220
Population Genetic Test Statistics
Pi
219.350953
Theta
187.543097
Tajima's D
1.006773
CLR
0.000316
CSRT
0.661866906654667
Interpretation
Uncertain
