Gene
KWMTBOMO14685
Pre Gene Modal
BGIBMGA009579
Annotation
PREDICTED:_pyridoxal_phosphate_phosphatase_PHOSPHO2-like_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 2.482
Sequence
CDS
ATGGCTTCTGTTGCTTTCTTTGATTTTGATCGTACAATAGTTGATGATGATTCTGATGCCACGATAATTAATAAATTAAGAGAGAAAAAGCCCCCGCCTGACTGGGAGAGCAGTAACCACGACTGGACTCCGTACATGAGTGATGTATTTGAGCACGCATACTCAGCCGGTCTAAGCGAAGAGGACATCCTGTCGTGCATAGCGTCAATGAAACCGAACCTCGGGGTGCAGCAGTTGATCAGGACGCTGTCCCAACAGGGCTGGGAGATTGTGGTCATCACAGACGCGAATAGTGTCTTCGTCAATCATTGGCTGACGGAGCACGGGTTGTTGCAATATATAACGAATGTCATCACAAATCGTGCGTTCTGGAGGCACAGCCGTCTGTACATCGAGCCCTATATGAAGCAGACGTCCTGCCCCAGGTGCCCTAAGAACCTCTGCAAGAGCGAGGCCCTGAGACGCTGGTGCTCGGAGATGCCGGCCAGGCGTATCGTGTATGTAGGGGACGGGAGAAACGATTACTGCCCCGCCACGTCGCTGCCCCCACACGCGACAGTGTTCCCCCGCCGCGGCTATCCACTGGACGACCTGGTGAAGAAGACGCTCTCGTCTCCCAACCCTCAAGTGAAGGCCAAAGTGATACCCTGGGACAACTGCTACAGGATACTCCAGGAACTCTACCCGGACATCATGGTGGATAATTAG
Protein
MASVAFFDFDRTIVDDDSDATIINKLREKKPPPDWESSNHDWTPYMSDVFEHAYSAGLSEEDILSCIASMKPNLGVQQLIRTLSQQGWEIVVITDANSVFVNHWLTEHGLLQYITNVITNRAFWRHSRLYIEPYMKQTSCPRCPKNLCKSEALRRWCSEMPARRIVYVGDGRNDYCPATSLPPHATVFPRRGYPLDDLVKKTLSSPNPQVKAKVIPWDNCYRILQELYPDIMVDN
Summary
Cofactor
Mg(2+)
Uniprot
H9JJ78
A0A194QFI6
A0A0L7KSZ1
A0A2A4JRX0
A0A212FBW3
A0A2H1VML4
+ More
A0A2W1BHF5 A0A2P2I3B9 A0A2H8TVU7 E9G2T8 A0A0P5UF79 A0A162P635 A0A0P6ITD6 J3JYP1 A0A0P4XE87 N6UE28 A0A0P4VNS8 A0A3Q3KR18 A0A2U9CRV8 A0A3P9QFU7 A0A3B5BG61 U4UFM8 A0A2A3EB76 A0A3Q3EJ82 A0A3Q3NB98 A0A088A276 A0A2S2PJ61 A0A3B5LCF9 A0A3Q1G1Q8 A0A3B3ZXX6 A0A087YQK8 A0A3Q3MLW1 A0A3Q0RHK5 A0A2I4CDN7 I3KXX3 A0A3Q2Y961 A0A3B3Z081 C4WW78 J9K4I9 G3P3B8 A0A3Q1D061 A0A3Q3QV54 A0A0A9WZ77 A0A0S7HAZ4 A0A3Q1H0T8 A0A154NW46 A0A3P8WNL4 H2UYE6 A0A3B3U7I2 A0A3Q1JML6 A0A2J7PJC8 A0A0K8TH61 A0A0A9X9L4 A0A0P4W1U5 A0A3B4WI69 A0A0A9X299 A0A3Q2CX20 A0A3B4VPF3 M4AZ91 A0A154MKD9 R7VHD7 A0A3P8RY01 H2LD81 A0A3B3VGX4 A0A3B3WMG5 A0A1A8UDE6 A0A087YPW4 A0A3B4G3R5 A0A3Q2W6W7 A0A3P8NB51 A0A3P9D830 A0A3Q3XKJ5 A0A3P8VAU9 A0A1A7YG39 A0A1A8IXG1 A0A1A8A511 A0A1A8BT90 A0A1A8Q469 I3KYN6 M4AZL9 A0A0S7H697 A0A3P9NS49 A0A3B5MHQ9 C1BR97 A0A1A8N758 A0A1A7WGE7 A0A3P9KKB1 A0A1A8LGF0 A0A3P9H204 A0A195B0A7 A0A158NKU6 A0A146W466 A0A3Q2QMZ8 A0A3B4VG10 A0A1B6M9F2 A0A3Q4HYB8 A0A0C9RDB3
A0A2W1BHF5 A0A2P2I3B9 A0A2H8TVU7 E9G2T8 A0A0P5UF79 A0A162P635 A0A0P6ITD6 J3JYP1 A0A0P4XE87 N6UE28 A0A0P4VNS8 A0A3Q3KR18 A0A2U9CRV8 A0A3P9QFU7 A0A3B5BG61 U4UFM8 A0A2A3EB76 A0A3Q3EJ82 A0A3Q3NB98 A0A088A276 A0A2S2PJ61 A0A3B5LCF9 A0A3Q1G1Q8 A0A3B3ZXX6 A0A087YQK8 A0A3Q3MLW1 A0A3Q0RHK5 A0A2I4CDN7 I3KXX3 A0A3Q2Y961 A0A3B3Z081 C4WW78 J9K4I9 G3P3B8 A0A3Q1D061 A0A3Q3QV54 A0A0A9WZ77 A0A0S7HAZ4 A0A3Q1H0T8 A0A154NW46 A0A3P8WNL4 H2UYE6 A0A3B3U7I2 A0A3Q1JML6 A0A2J7PJC8 A0A0K8TH61 A0A0A9X9L4 A0A0P4W1U5 A0A3B4WI69 A0A0A9X299 A0A3Q2CX20 A0A3B4VPF3 M4AZ91 A0A154MKD9 R7VHD7 A0A3P8RY01 H2LD81 A0A3B3VGX4 A0A3B3WMG5 A0A1A8UDE6 A0A087YPW4 A0A3B4G3R5 A0A3Q2W6W7 A0A3P8NB51 A0A3P9D830 A0A3Q3XKJ5 A0A3P8VAU9 A0A1A7YG39 A0A1A8IXG1 A0A1A8A511 A0A1A8BT90 A0A1A8Q469 I3KYN6 M4AZL9 A0A0S7H697 A0A3P9NS49 A0A3B5MHQ9 C1BR97 A0A1A8N758 A0A1A7WGE7 A0A3P9KKB1 A0A1A8LGF0 A0A3P9H204 A0A195B0A7 A0A158NKU6 A0A146W466 A0A3Q2QMZ8 A0A3B4VG10 A0A1B6M9F2 A0A3Q4HYB8 A0A0C9RDB3
Pubmed
EMBL
BABH01036416
BABH01036417
KQ459249
KPJ02231.1
JTDY01006265
KOB66161.1
+ More
NWSH01000762 PCG74364.1 AGBW02009261 OWR51236.1 ODYU01003372 SOQ42047.1 KZ150125 PZC73124.1 IACF01002726 LAB68370.1 GFXV01005867 MBW17672.1 GL732530 EFX86450.1 GDIP01113861 JAL89853.1 LRGB01000512 KZS18554.1 GDIQ01000384 JAN94353.1 BT128371 AEE63329.1 GDIP01244410 JAI78991.1 APGK01026301 KB740624 ENN79965.1 GDKW01003189 JAI53406.1 CP026261 AWP19328.1 KB632335 ERL92789.1 KZ288311 PBC28459.1 GGMR01016870 MBY29489.1 AYCK01015888 AERX01007407 AK341906 BAH72148.1 ABLF02026088 GBHO01029817 GBHO01029812 GBRD01006108 JAG13787.1 JAG13792.1 JAG59713.1 GBYX01444761 JAO36659.1 KQ434772 KZC03915.1 NEVH01024949 PNF16441.1 GBRD01000893 JAG64928.1 GBHO01029814 JAG13790.1 GDRN01079587 JAI62371.1 GBHO01029813 JAG13791.1 GCES01062505 JAR23818.1 AMQN01003891 KB292163 ELU18039.1 HAEJ01004875 SBS45332.1 AYCK01007245 HADX01006887 SBP29119.1 HAED01015577 HAEE01009884 SBR02022.1 HADY01011722 SBP50207.1 HADZ01005799 HAEA01002026 SBP69740.1 HAEI01004392 SBR88580.1 AERX01007582 GBYX01444760 JAO36660.1 BT077126 ACO11550.1 HAEH01000447 SBR64908.1 HADW01003204 SBP04604.1 HAEF01006295 HAEG01004600 SBR43677.1 KQ976692 KYM77891.1 ADTU01018871 GCES01062504 JAR23819.1 GEBQ01007414 JAT32563.1 GBYB01014579 JAG84346.1
NWSH01000762 PCG74364.1 AGBW02009261 OWR51236.1 ODYU01003372 SOQ42047.1 KZ150125 PZC73124.1 IACF01002726 LAB68370.1 GFXV01005867 MBW17672.1 GL732530 EFX86450.1 GDIP01113861 JAL89853.1 LRGB01000512 KZS18554.1 GDIQ01000384 JAN94353.1 BT128371 AEE63329.1 GDIP01244410 JAI78991.1 APGK01026301 KB740624 ENN79965.1 GDKW01003189 JAI53406.1 CP026261 AWP19328.1 KB632335 ERL92789.1 KZ288311 PBC28459.1 GGMR01016870 MBY29489.1 AYCK01015888 AERX01007407 AK341906 BAH72148.1 ABLF02026088 GBHO01029817 GBHO01029812 GBRD01006108 JAG13787.1 JAG13792.1 JAG59713.1 GBYX01444761 JAO36659.1 KQ434772 KZC03915.1 NEVH01024949 PNF16441.1 GBRD01000893 JAG64928.1 GBHO01029814 JAG13790.1 GDRN01079587 JAI62371.1 GBHO01029813 JAG13791.1 GCES01062505 JAR23818.1 AMQN01003891 KB292163 ELU18039.1 HAEJ01004875 SBS45332.1 AYCK01007245 HADX01006887 SBP29119.1 HAED01015577 HAEE01009884 SBR02022.1 HADY01011722 SBP50207.1 HADZ01005799 HAEA01002026 SBP69740.1 HAEI01004392 SBR88580.1 AERX01007582 GBYX01444760 JAO36660.1 BT077126 ACO11550.1 HAEH01000447 SBR64908.1 HADW01003204 SBP04604.1 HAEF01006295 HAEG01004600 SBR43677.1 KQ976692 KYM77891.1 ADTU01018871 GCES01062504 JAR23819.1 GEBQ01007414 JAT32563.1 GBYB01014579 JAG84346.1
Proteomes
UP000005204
UP000053268
UP000037510
UP000218220
UP000007151
UP000000305
+ More
UP000076858 UP000019118 UP000261600 UP000246464 UP000242638 UP000261400 UP000030742 UP000242457 UP000261660 UP000261640 UP000005203 UP000261380 UP000257200 UP000261520 UP000028760 UP000261340 UP000192220 UP000005207 UP000264820 UP000261480 UP000007819 UP000007635 UP000257160 UP000265040 UP000076502 UP000265120 UP000005226 UP000261500 UP000235965 UP000261360 UP000265020 UP000261420 UP000002852 UP000014760 UP000265080 UP000001038 UP000261460 UP000264840 UP000265100 UP000265160 UP000261620 UP000265180 UP000265200 UP000078540 UP000005205 UP000265000 UP000261580
UP000076858 UP000019118 UP000261600 UP000246464 UP000242638 UP000261400 UP000030742 UP000242457 UP000261660 UP000261640 UP000005203 UP000261380 UP000257200 UP000261520 UP000028760 UP000261340 UP000192220 UP000005207 UP000264820 UP000261480 UP000007819 UP000007635 UP000257160 UP000265040 UP000076502 UP000265120 UP000005226 UP000261500 UP000235965 UP000261360 UP000265020 UP000261420 UP000002852 UP000014760 UP000265080 UP000001038 UP000261460 UP000264840 UP000265100 UP000265160 UP000261620 UP000265180 UP000265200 UP000078540 UP000005205 UP000265000 UP000261580
PRIDE
Pfam
PF06888 Put_Phosphatase
Interpro
SUPFAM
SSF56784
SSF56784
Gene 3D
ProteinModelPortal
H9JJ78
A0A194QFI6
A0A0L7KSZ1
A0A2A4JRX0
A0A212FBW3
A0A2H1VML4
+ More
A0A2W1BHF5 A0A2P2I3B9 A0A2H8TVU7 E9G2T8 A0A0P5UF79 A0A162P635 A0A0P6ITD6 J3JYP1 A0A0P4XE87 N6UE28 A0A0P4VNS8 A0A3Q3KR18 A0A2U9CRV8 A0A3P9QFU7 A0A3B5BG61 U4UFM8 A0A2A3EB76 A0A3Q3EJ82 A0A3Q3NB98 A0A088A276 A0A2S2PJ61 A0A3B5LCF9 A0A3Q1G1Q8 A0A3B3ZXX6 A0A087YQK8 A0A3Q3MLW1 A0A3Q0RHK5 A0A2I4CDN7 I3KXX3 A0A3Q2Y961 A0A3B3Z081 C4WW78 J9K4I9 G3P3B8 A0A3Q1D061 A0A3Q3QV54 A0A0A9WZ77 A0A0S7HAZ4 A0A3Q1H0T8 A0A154NW46 A0A3P8WNL4 H2UYE6 A0A3B3U7I2 A0A3Q1JML6 A0A2J7PJC8 A0A0K8TH61 A0A0A9X9L4 A0A0P4W1U5 A0A3B4WI69 A0A0A9X299 A0A3Q2CX20 A0A3B4VPF3 M4AZ91 A0A154MKD9 R7VHD7 A0A3P8RY01 H2LD81 A0A3B3VGX4 A0A3B3WMG5 A0A1A8UDE6 A0A087YPW4 A0A3B4G3R5 A0A3Q2W6W7 A0A3P8NB51 A0A3P9D830 A0A3Q3XKJ5 A0A3P8VAU9 A0A1A7YG39 A0A1A8IXG1 A0A1A8A511 A0A1A8BT90 A0A1A8Q469 I3KYN6 M4AZL9 A0A0S7H697 A0A3P9NS49 A0A3B5MHQ9 C1BR97 A0A1A8N758 A0A1A7WGE7 A0A3P9KKB1 A0A1A8LGF0 A0A3P9H204 A0A195B0A7 A0A158NKU6 A0A146W466 A0A3Q2QMZ8 A0A3B4VG10 A0A1B6M9F2 A0A3Q4HYB8 A0A0C9RDB3
A0A2W1BHF5 A0A2P2I3B9 A0A2H8TVU7 E9G2T8 A0A0P5UF79 A0A162P635 A0A0P6ITD6 J3JYP1 A0A0P4XE87 N6UE28 A0A0P4VNS8 A0A3Q3KR18 A0A2U9CRV8 A0A3P9QFU7 A0A3B5BG61 U4UFM8 A0A2A3EB76 A0A3Q3EJ82 A0A3Q3NB98 A0A088A276 A0A2S2PJ61 A0A3B5LCF9 A0A3Q1G1Q8 A0A3B3ZXX6 A0A087YQK8 A0A3Q3MLW1 A0A3Q0RHK5 A0A2I4CDN7 I3KXX3 A0A3Q2Y961 A0A3B3Z081 C4WW78 J9K4I9 G3P3B8 A0A3Q1D061 A0A3Q3QV54 A0A0A9WZ77 A0A0S7HAZ4 A0A3Q1H0T8 A0A154NW46 A0A3P8WNL4 H2UYE6 A0A3B3U7I2 A0A3Q1JML6 A0A2J7PJC8 A0A0K8TH61 A0A0A9X9L4 A0A0P4W1U5 A0A3B4WI69 A0A0A9X299 A0A3Q2CX20 A0A3B4VPF3 M4AZ91 A0A154MKD9 R7VHD7 A0A3P8RY01 H2LD81 A0A3B3VGX4 A0A3B3WMG5 A0A1A8UDE6 A0A087YPW4 A0A3B4G3R5 A0A3Q2W6W7 A0A3P8NB51 A0A3P9D830 A0A3Q3XKJ5 A0A3P8VAU9 A0A1A7YG39 A0A1A8IXG1 A0A1A8A511 A0A1A8BT90 A0A1A8Q469 I3KYN6 M4AZL9 A0A0S7H697 A0A3P9NS49 A0A3B5MHQ9 C1BR97 A0A1A8N758 A0A1A7WGE7 A0A3P9KKB1 A0A1A8LGF0 A0A3P9H204 A0A195B0A7 A0A158NKU6 A0A146W466 A0A3Q2QMZ8 A0A3B4VG10 A0A1B6M9F2 A0A3Q4HYB8 A0A0C9RDB3
Ontologies
PATHWAY
GO
PANTHER
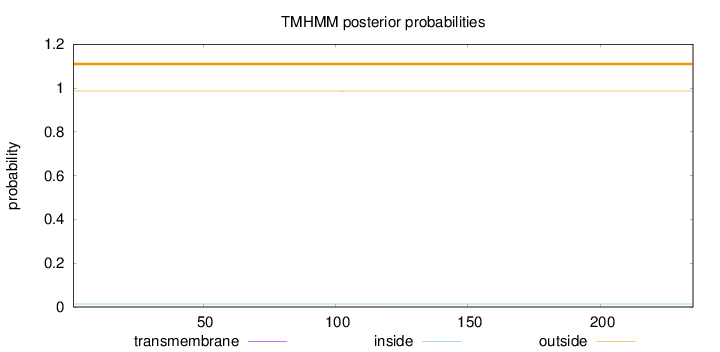
Topology
Length:
235
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0042
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01376
outside
1 - 235
Population Genetic Test Statistics
Pi
150.874172
Theta
135.939315
Tajima's D
0.134013
CLR
29.117215
CSRT
0.401229938503075
Interpretation
Uncertain
