Gene
KWMTBOMO14684
Pre Gene Modal
BGIBMGA009617
Annotation
putative_sorting_nexin_family_member_30_[Operophtera_brumata]
Full name
Sorting nexin-30
Location in the cell
Nuclear Reliability : 3.136
Sequence
CDS
ATGGCCAGTGAAAGCAGTTCAGCAATGCTGGACATGACAGATGCTGACTCCGACGATATGGACATGCCGACTCTCATGGACGATCAGGAGCCATCACCGCTCATCATGGAGCACGGCTATGACATTGCAATTAAGGTCGATGCCCCCATAAAACAGCTGTCGACCCTCGAGACGTATGTGACGTACCGGGTGCGTTGCGTGTGCGCGCGGTGGCCGACCCCGCCCAGCGTGAGACGACGGTACAATCACTTTAAGGTGCTGCACAAGCGGCTGTCGGCGGCGCACCCCCTCACCGCGGCGCCCCCCGCGCCCCCGCTGCACTCCGCGCGCCAGCAGCTCGACAGATACTCGGCGTCGTTTGTCGCCATAAGGACCTTAGCCCTCAACGCGTTTCTAGATCGCATCGCCAAACATCCGATCCTGACACACAGCGAAGACTTCAGATTGTTTCTGACCACGCCAGATGAGGAAATCGATAAGGTGTTCCGGAGTGAGAATAACGCGTTGAATCTGTGGGGGCTGGGCAGCGCGCTAGCGTACGGTGGGGACGGGCGTCACTCCAACGGAGTCCGAGTAAAAGATCCAGAGTTTTCATCGGCAGTCGACTACCTGAACGCTCTACAAGACAAGCTGGCCACCCTCTGTGATCTGACCACGAAACTATACAGGGGGACCGCCGCCCTCGCCGCCGAGTACTCGAGCATGCAGCGCGTGTGCGACGTGTGGTCGACGCAGTGCGGGGCGCGGTGCGGGGGAACGCAGCGGGGCGTGCGGGGGGTGTCGGCGGGCGCGGGGGCGGCGGGGCAGGCGGCGGGGGGGCGGCCCTCCCCCTCCCACCGCGCCGTGCTGCCCCTGCTGGCCGCCTACGCCGCGGCCCAGGCCGATAAGATAAGGCAGCGGGACAGGATGCACGCGGAATACGTGTCTGGGACGGACACCAGCGACGACCTGCAGAACAGAGTCGAGGAGGCGACCGAGGCTCTGCGCAGTGAGCTCTGGTCGTGGACCGCCAAGACCCATCAGGACATCAAGGGCGTCCTGGCGTACATAGCCACCAGACAGGTGTCGGTGCTAGCGCAGTCGCTGCAGGGCTGGGAGCAGGCGCTCCGCGTGGCCACCGACACGAGCGTCCACCATTTGTTCCAGACGGTCTCCAAGAATGGCGTCCGCAACTTGTCCCCCACCACGTCGGCGGCCCCCCACGCGTCCCCCCAAGCGGACCCCCTGCGCCCCGACTCGCCCGACGGCACGGACACCTAA
Protein
MASESSSAMLDMTDADSDDMDMPTLMDDQEPSPLIMEHGYDIAIKVDAPIKQLSTLETYVTYRVRCVCARWPTPPSVRRRYNHFKVLHKRLSAAHPLTAAPPAPPLHSARQQLDRYSASFVAIRTLALNAFLDRIAKHPILTHSEDFRLFLTTPDEEIDKVFRSENNALNLWGLGSALAYGGDGRHSNGVRVKDPEFSSAVDYLNALQDKLATLCDLTTKLYRGTAALAAEYSSMQRVCDVWSTQCGARCGGTQRGVRGVSAGAGAAGQAAGGRPSPSHRAVLPLLAAYAAAQADKIRQRDRMHAEYVSGTDTSDDLQNRVEEATEALRSELWSWTAKTHQDIKGVLAYIATRQVSVLAQSLQGWEQALRVATDTSVHHLFQTVSKNGVRNLSPTTSAAPHASPQADPLRPDSPDGTDT
Summary
Description
May be involved in several stages of intracellular trafficking.
Similarity
Belongs to the sorting nexin family.
Keywords
Protein transport
Transport
Complete proteome
Phosphoprotein
Polymorphism
Reference proteome
Feature
chain Sorting nexin-30
sequence variant In dbSNP:rs2796036.
sequence variant In dbSNP:rs2796036.
Uniprot
A0A2H1VFI5
A0A2A4JRB6
A0A2W1BIR7
A0A3S2N724
A0A0L7LP91
A0A3B3R040
+ More
A0A3B3R050 J3S4R7 A0A151NEA0 A0A3B3QZG4 G1PK69 A0A151NED2 R7UKX8 Q6GMJ0 H2PT32 A0A2I3GAI5 A0A2K5HPI8 A0A1U8DGR0 W5M4S0 A0A2D4PC24 A0A2K6MAK8 A0A0R4IIY5 A0A3B4E080 A0A2K6PSP1 H0YSM4 A0A2D4PEK8 D4A060 A0A2D4JYZ4 U3JBH0 A0A218UUH9 G7NG62 G1KTU9 A0A2K5RVJ8 K7FWP8 A0A3Q7X5H3 Q4V7P7 A0A3Q0E991 F1SNA5 A0A2R9AWL4 A0A2K5MU48 G3QNL2 K7A3S9 Q5VWJ9 M3W6K4 A0A091N516 A0A2U3VPI0 A0A2D4LQH3 A0A2K5YNA8 A0A093GH97 A0A2Y9H609 A0A2U3YVH6 A0A2D4N287 F6YJC1 A0A3Q7QE99 A0A2K6C194 A0A2K5TWJ5 A0A096P0U9 F7GDT2 F6YJC8 A0A0D9RIV4 A0A2K5D4E2 A0A2K6UVH1 A0A2Y9JAR9 M3Y8M5 G3T2I5 U3DYT1 A0A093RI49 A0A091I1L3 U3F1I2 A0A2K5YNC1 U6D9B4 A0A3Q7SPC0 A0A384DGX4 R7VRS9 A0A2I0M5G9 A0A091R0J6 A0A2I0U8B8 A0A087R110 A0A093HI11 A0A1U7QSI0 A0A093L797 A0A091U1B5 A0A091LB03 A0A0A0ARP1
A0A3B3R050 J3S4R7 A0A151NEA0 A0A3B3QZG4 G1PK69 A0A151NED2 R7UKX8 Q6GMJ0 H2PT32 A0A2I3GAI5 A0A2K5HPI8 A0A1U8DGR0 W5M4S0 A0A2D4PC24 A0A2K6MAK8 A0A0R4IIY5 A0A3B4E080 A0A2K6PSP1 H0YSM4 A0A2D4PEK8 D4A060 A0A2D4JYZ4 U3JBH0 A0A218UUH9 G7NG62 G1KTU9 A0A2K5RVJ8 K7FWP8 A0A3Q7X5H3 Q4V7P7 A0A3Q0E991 F1SNA5 A0A2R9AWL4 A0A2K5MU48 G3QNL2 K7A3S9 Q5VWJ9 M3W6K4 A0A091N516 A0A2U3VPI0 A0A2D4LQH3 A0A2K5YNA8 A0A093GH97 A0A2Y9H609 A0A2U3YVH6 A0A2D4N287 F6YJC1 A0A3Q7QE99 A0A2K6C194 A0A2K5TWJ5 A0A096P0U9 F7GDT2 F6YJC8 A0A0D9RIV4 A0A2K5D4E2 A0A2K6UVH1 A0A2Y9JAR9 M3Y8M5 G3T2I5 U3DYT1 A0A093RI49 A0A091I1L3 U3F1I2 A0A2K5YNC1 U6D9B4 A0A3Q7SPC0 A0A384DGX4 R7VRS9 A0A2I0M5G9 A0A091R0J6 A0A2I0U8B8 A0A087R110 A0A093HI11 A0A1U7QSI0 A0A093L797 A0A091U1B5 A0A091LB03 A0A0A0ARP1
Pubmed
EMBL
ODYU01002303
SOQ39620.1
NWSH01000762
PCG74359.1
KZ150125
PZC73127.1
+ More
RSAL01000277 RVE43093.1 JTDY01000437 KOB77169.1 JU175238 AFJ50762.1 AKHW03003207 KYO35143.1 AAPE02049003 AAPE02049004 AAPE02049005 KYO35142.1 AMQN01007257 KB300307 ELU06895.1 BC074057 AAH74057.1 ABGA01378870 ABGA01378871 ABGA01378872 ABGA01378873 ABGA01378874 ABGA01378875 ABGA01378876 ABGA01378877 ABGA01378878 ABGA01378879 NDHI03003380 PNJ72517.1 ADFV01019658 ADFV01019659 ADFV01019660 ADFV01019661 ADFV01019662 ADFV01019663 ADFV01019664 ADFV01019665 ADFV01019666 ADFV01019667 AHAT01012601 AHAT01012602 IACN01060121 LAB55552.1 CU929325 ABQF01045193 ABQF01045194 ABQF01045195 ABQF01045196 ABQF01045197 IACN01060119 LAB55546.1 AABR07048403 AABR07048404 AABR07048405 CH474039 EDL91629.1 IACL01018701 LAB01658.1 AGTO01011559 MUZQ01000127 OWK57286.1 CM001267 EHH23861.1 AGCU01164325 AGCU01164326 AGCU01164327 AGCU01164328 BC097784 AEMK02000005 AJFE02033618 AJFE02033619 AJFE02033620 AJFE02033621 AJFE02033622 AJFE02033623 AJFE02033624 CABD030065920 CABD030065921 GABC01009095 GABF01009109 GABD01010502 NBAG03000559 JAA02243.1 JAA13036.1 JAA22598.1 PNI15363.1 AL390067 AL360235 AL139041 AANG04004382 KK842725 KFP84893.1 IACM01032402 LAB23264.1 KL216234 KFV68598.1 IACM01135356 LAB39103.1 AQIA01023754 AQIA01023755 AQIA01023756 AQIA01023757 AHZZ02008279 AHZZ02008280 AHZZ02008281 JSUE03014429 JSUE03014430 JSUE03014431 JSUE03014432 JSUE03014433 AQIB01022376 AQIB01022377 AQIB01022378 AQIB01022379 AQIB01022380 AEYP01022963 AEYP01022964 AEYP01022965 AEYP01022966 GAMS01004645 JAB18491.1 KL225291 KFW70579.1 KL218126 KFP02384.1 GAMR01007809 GAMP01001579 GAMP01001578 JAB26123.1 JAB51177.1 HAAF01007731 CCP79555.1 KB376033 EMC78465.1 AKCR02000037 PKK24921.1 KK709966 KFQ32714.1 KZ506009 PKU42295.1 KL226038 KFM07164.1 KL206418 KFV82288.1 KK599163 KFW05126.1 KK410225 KFQ83852.1 KL313394 KFP53534.1 KL872964 KGL97244.1
RSAL01000277 RVE43093.1 JTDY01000437 KOB77169.1 JU175238 AFJ50762.1 AKHW03003207 KYO35143.1 AAPE02049003 AAPE02049004 AAPE02049005 KYO35142.1 AMQN01007257 KB300307 ELU06895.1 BC074057 AAH74057.1 ABGA01378870 ABGA01378871 ABGA01378872 ABGA01378873 ABGA01378874 ABGA01378875 ABGA01378876 ABGA01378877 ABGA01378878 ABGA01378879 NDHI03003380 PNJ72517.1 ADFV01019658 ADFV01019659 ADFV01019660 ADFV01019661 ADFV01019662 ADFV01019663 ADFV01019664 ADFV01019665 ADFV01019666 ADFV01019667 AHAT01012601 AHAT01012602 IACN01060121 LAB55552.1 CU929325 ABQF01045193 ABQF01045194 ABQF01045195 ABQF01045196 ABQF01045197 IACN01060119 LAB55546.1 AABR07048403 AABR07048404 AABR07048405 CH474039 EDL91629.1 IACL01018701 LAB01658.1 AGTO01011559 MUZQ01000127 OWK57286.1 CM001267 EHH23861.1 AGCU01164325 AGCU01164326 AGCU01164327 AGCU01164328 BC097784 AEMK02000005 AJFE02033618 AJFE02033619 AJFE02033620 AJFE02033621 AJFE02033622 AJFE02033623 AJFE02033624 CABD030065920 CABD030065921 GABC01009095 GABF01009109 GABD01010502 NBAG03000559 JAA02243.1 JAA13036.1 JAA22598.1 PNI15363.1 AL390067 AL360235 AL139041 AANG04004382 KK842725 KFP84893.1 IACM01032402 LAB23264.1 KL216234 KFV68598.1 IACM01135356 LAB39103.1 AQIA01023754 AQIA01023755 AQIA01023756 AQIA01023757 AHZZ02008279 AHZZ02008280 AHZZ02008281 JSUE03014429 JSUE03014430 JSUE03014431 JSUE03014432 JSUE03014433 AQIB01022376 AQIB01022377 AQIB01022378 AQIB01022379 AQIB01022380 AEYP01022963 AEYP01022964 AEYP01022965 AEYP01022966 GAMS01004645 JAB18491.1 KL225291 KFW70579.1 KL218126 KFP02384.1 GAMR01007809 GAMP01001579 GAMP01001578 JAB26123.1 JAB51177.1 HAAF01007731 CCP79555.1 KB376033 EMC78465.1 AKCR02000037 PKK24921.1 KK709966 KFQ32714.1 KZ506009 PKU42295.1 KL226038 KFM07164.1 KL206418 KFV82288.1 KK599163 KFW05126.1 KK410225 KFQ83852.1 KL313394 KFP53534.1 KL872964 KGL97244.1
Proteomes
UP000218220
UP000283053
UP000037510
UP000261540
UP000050525
UP000001074
+ More
UP000014760 UP000001595 UP000001073 UP000233080 UP000189705 UP000018468 UP000233180 UP000000437 UP000261440 UP000233200 UP000007754 UP000002494 UP000016665 UP000197619 UP000001646 UP000233040 UP000007267 UP000286642 UP000189704 UP000008227 UP000240080 UP000233060 UP000001519 UP000005640 UP000011712 UP000245340 UP000233140 UP000053875 UP000248481 UP000245341 UP000002279 UP000286641 UP000233120 UP000233100 UP000028761 UP000006718 UP000029965 UP000233020 UP000233220 UP000248482 UP000000715 UP000007646 UP000054081 UP000054308 UP000008225 UP000286640 UP000261680 UP000053872 UP000053286 UP000053584 UP000189706 UP000053858
UP000014760 UP000001595 UP000001073 UP000233080 UP000189705 UP000018468 UP000233180 UP000000437 UP000261440 UP000233200 UP000007754 UP000002494 UP000016665 UP000197619 UP000001646 UP000233040 UP000007267 UP000286642 UP000189704 UP000008227 UP000240080 UP000233060 UP000001519 UP000005640 UP000011712 UP000245340 UP000233140 UP000053875 UP000248481 UP000245341 UP000002279 UP000286641 UP000233120 UP000233100 UP000028761 UP000006718 UP000029965 UP000233020 UP000233220 UP000248482 UP000000715 UP000007646 UP000054081 UP000054308 UP000008225 UP000286640 UP000261680 UP000053872 UP000053286 UP000053584 UP000189706 UP000053858
Interpro
Gene 3D
ProteinModelPortal
A0A2H1VFI5
A0A2A4JRB6
A0A2W1BIR7
A0A3S2N724
A0A0L7LP91
A0A3B3R040
+ More
A0A3B3R050 J3S4R7 A0A151NEA0 A0A3B3QZG4 G1PK69 A0A151NED2 R7UKX8 Q6GMJ0 H2PT32 A0A2I3GAI5 A0A2K5HPI8 A0A1U8DGR0 W5M4S0 A0A2D4PC24 A0A2K6MAK8 A0A0R4IIY5 A0A3B4E080 A0A2K6PSP1 H0YSM4 A0A2D4PEK8 D4A060 A0A2D4JYZ4 U3JBH0 A0A218UUH9 G7NG62 G1KTU9 A0A2K5RVJ8 K7FWP8 A0A3Q7X5H3 Q4V7P7 A0A3Q0E991 F1SNA5 A0A2R9AWL4 A0A2K5MU48 G3QNL2 K7A3S9 Q5VWJ9 M3W6K4 A0A091N516 A0A2U3VPI0 A0A2D4LQH3 A0A2K5YNA8 A0A093GH97 A0A2Y9H609 A0A2U3YVH6 A0A2D4N287 F6YJC1 A0A3Q7QE99 A0A2K6C194 A0A2K5TWJ5 A0A096P0U9 F7GDT2 F6YJC8 A0A0D9RIV4 A0A2K5D4E2 A0A2K6UVH1 A0A2Y9JAR9 M3Y8M5 G3T2I5 U3DYT1 A0A093RI49 A0A091I1L3 U3F1I2 A0A2K5YNC1 U6D9B4 A0A3Q7SPC0 A0A384DGX4 R7VRS9 A0A2I0M5G9 A0A091R0J6 A0A2I0U8B8 A0A087R110 A0A093HI11 A0A1U7QSI0 A0A093L797 A0A091U1B5 A0A091LB03 A0A0A0ARP1
A0A3B3R050 J3S4R7 A0A151NEA0 A0A3B3QZG4 G1PK69 A0A151NED2 R7UKX8 Q6GMJ0 H2PT32 A0A2I3GAI5 A0A2K5HPI8 A0A1U8DGR0 W5M4S0 A0A2D4PC24 A0A2K6MAK8 A0A0R4IIY5 A0A3B4E080 A0A2K6PSP1 H0YSM4 A0A2D4PEK8 D4A060 A0A2D4JYZ4 U3JBH0 A0A218UUH9 G7NG62 G1KTU9 A0A2K5RVJ8 K7FWP8 A0A3Q7X5H3 Q4V7P7 A0A3Q0E991 F1SNA5 A0A2R9AWL4 A0A2K5MU48 G3QNL2 K7A3S9 Q5VWJ9 M3W6K4 A0A091N516 A0A2U3VPI0 A0A2D4LQH3 A0A2K5YNA8 A0A093GH97 A0A2Y9H609 A0A2U3YVH6 A0A2D4N287 F6YJC1 A0A3Q7QE99 A0A2K6C194 A0A2K5TWJ5 A0A096P0U9 F7GDT2 F6YJC8 A0A0D9RIV4 A0A2K5D4E2 A0A2K6UVH1 A0A2Y9JAR9 M3Y8M5 G3T2I5 U3DYT1 A0A093RI49 A0A091I1L3 U3F1I2 A0A2K5YNC1 U6D9B4 A0A3Q7SPC0 A0A384DGX4 R7VRS9 A0A2I0M5G9 A0A091R0J6 A0A2I0U8B8 A0A087R110 A0A093HI11 A0A1U7QSI0 A0A093L797 A0A091U1B5 A0A091LB03 A0A0A0ARP1
PDB
3IQ2
E-value=6.178e-12,
Score=171
Ontologies
GO
PANTHER
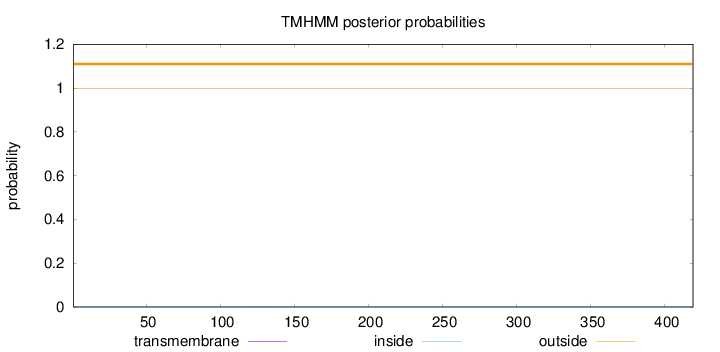
Topology
Length:
419
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00528
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00276
outside
1 - 419
Population Genetic Test Statistics
Pi
207.450745
Theta
155.915023
Tajima's D
1.27381
CLR
0.311041
CSRT
0.734513274336283
Interpretation
Uncertain
