Pre Gene Modal
BGIBMGA009612
Annotation
PREDICTED:_probable_proline--tRNA_ligase?_mitochondrial_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.446 Mitochondrial Reliability : 1.444
Sequence
CDS
ATGAGATATTTATCTCGTATATTTCAACCAGTTATAACAATACCAAAGAATGCTAAGATAAAGAACACGGAAATAACATGTAAAAGTCAGAAGCTGTTACTAGAATGTGGTTTAGTCCGCCCAACTACGTCTGGCTTCTTCACTCTTCTCCCCTTGGCACGTAGAGCACTGGATAAACTTGAGAACCTCATTAAAACCTGCATAGAATCTTCTGGAGCGCAAAGATTGTCATTGCCGGCTCTCACGTCAGCGGGGCTGTGGGCGAAGACTGGCCGCCTGGAGAATGCTGCTGCAGAGCTATTGACCCTGGAAGATAGACACAACAAAAAATATCTGTTGGCTCCGACATACGAAGAGGCAATCGCGGAGTTATTGTCAGACCTCAAGCCGATCTCATACAAGCAACTACCGATTATATTATATCAGGTAAAATTAAACTGTTCTTAG
Protein
MRYLSRIFQPVITIPKNAKIKNTEITCKSQKLLLECGLVRPTTSGFFTLLPLARRALDKLENLIKTCIESSGAQRLSLPALTSAGLWAKTGRLENAAAELLTLEDRHNKKYLLAPTYEEAIAELLSDLKPISYKQLPIILYQVKLNCS
Summary
Uniprot
H9JJB1
A0A2A4JKU7
A0A2H1WFD1
A0A2W1BJX6
S4P313
A0A194Q9P0
+ More
A0A212EIR1 A0A194QUG2 A0A0A1XAV5 A0A084VH98 A0A2P8ZFE6 A0A034VBT6 A0A0K8UEV5 A0A0K8V8I2 A0A336MAS1 A0A0K8UCF6 A0A336M592 A0A1B6DP72 D6WC57 A0A1B6C484 A0A1B6HIW0 A0A182QU97 A0A1B6GM72 A0A1B6MFX8 Q0IEE6 A0A1I8MRT3 A0A182JII1 A0A1A9VHK3 A0A0M4EN92 A0A1I8JUZ4 A0A182V1E4 B4KYF2 A0A182G3B0 A0A1B0FBZ7 A0A1B0AG19 B4HTF1 A0A182KXJ1 Q7QAP8 A0A1B0BFR2 A0A182U533 A0A1I8JTP7 A0A2C9H8K8 A0A1A9Y6U0 A0A182Y7C6 A0A3B0KMU1 A0A1J1J937 A0A1Y1LL22 B4LDT6 A0A1B0C8D2 A0A1I8P2Z3 A0A182M4Y8 B4QNK7 Q9VZY9 B3M7I7 A0A1Y9IVS2 B4IZD4 B0XIT5 Q86P96 B3NBK6 A0A3R7PED5 Q29FB8 B4N398 A0A0P4W0H3 A0A0P4WKV3 A0A0P4WC38 A0A0P5P8E5 A0A1W4W297 B4PDC7 A0A1A9WFJ7 A0A0P5TR34 A0A1L8DHL5 A0A2H8TLW5 A0A0P5V1L7 A0A1L8DHM3 A0A0P5VHJ3 A0A067QQ20 A0A0A9YV78 A0A164Z717 A0A0P6I8D1 A0A0P5VYF4 A0A0P5R0S5 A0A0P5I0G0 A0A0P5WUP6 A0A0P5QKQ0 A0A0P5HAK0 A0A0P5AVD6 A0A0P5UNQ1 A0A0P5D1Q1 A0A2S2NSA3 A0A0P5XW94 A0A0P5SEW4 A0A0P6BPZ7 A0A0P5YND8 A0A0P5SQ69 A0A0P5X868 W5JJZ9 A0A182N4R5 W4XZ81 A0A182FNS6 A0A2S2Q7V4
A0A212EIR1 A0A194QUG2 A0A0A1XAV5 A0A084VH98 A0A2P8ZFE6 A0A034VBT6 A0A0K8UEV5 A0A0K8V8I2 A0A336MAS1 A0A0K8UCF6 A0A336M592 A0A1B6DP72 D6WC57 A0A1B6C484 A0A1B6HIW0 A0A182QU97 A0A1B6GM72 A0A1B6MFX8 Q0IEE6 A0A1I8MRT3 A0A182JII1 A0A1A9VHK3 A0A0M4EN92 A0A1I8JUZ4 A0A182V1E4 B4KYF2 A0A182G3B0 A0A1B0FBZ7 A0A1B0AG19 B4HTF1 A0A182KXJ1 Q7QAP8 A0A1B0BFR2 A0A182U533 A0A1I8JTP7 A0A2C9H8K8 A0A1A9Y6U0 A0A182Y7C6 A0A3B0KMU1 A0A1J1J937 A0A1Y1LL22 B4LDT6 A0A1B0C8D2 A0A1I8P2Z3 A0A182M4Y8 B4QNK7 Q9VZY9 B3M7I7 A0A1Y9IVS2 B4IZD4 B0XIT5 Q86P96 B3NBK6 A0A3R7PED5 Q29FB8 B4N398 A0A0P4W0H3 A0A0P4WKV3 A0A0P4WC38 A0A0P5P8E5 A0A1W4W297 B4PDC7 A0A1A9WFJ7 A0A0P5TR34 A0A1L8DHL5 A0A2H8TLW5 A0A0P5V1L7 A0A1L8DHM3 A0A0P5VHJ3 A0A067QQ20 A0A0A9YV78 A0A164Z717 A0A0P6I8D1 A0A0P5VYF4 A0A0P5R0S5 A0A0P5I0G0 A0A0P5WUP6 A0A0P5QKQ0 A0A0P5HAK0 A0A0P5AVD6 A0A0P5UNQ1 A0A0P5D1Q1 A0A2S2NSA3 A0A0P5XW94 A0A0P5SEW4 A0A0P6BPZ7 A0A0P5YND8 A0A0P5SQ69 A0A0P5X868 W5JJZ9 A0A182N4R5 W4XZ81 A0A182FNS6 A0A2S2Q7V4
Pubmed
19121390
28756777
23622113
26354079
22118469
25830018
+ More
24438588 29403074 25348373 18362917 19820115 17510324 25315136 17994087 26483478 20966253 12364791 14747013 17210077 25244985 28004739 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 17550304 24845553 25401762 26823975 20920257 23761445
24438588 29403074 25348373 18362917 19820115 17510324 25315136 17994087 26483478 20966253 12364791 14747013 17210077 25244985 28004739 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 17550304 24845553 25401762 26823975 20920257 23761445
EMBL
BABH01036404
NWSH01001085
PCG72705.1
ODYU01008302
SOQ51773.1
KZ150125
+ More
PZC73136.1 GAIX01006679 JAA85881.1 KQ459249 KPJ02223.1 AGBW02014604 OWR41350.1 KQ461108 KPJ08964.1 GBXI01006442 JAD07850.1 ATLV01013150 KE524842 KFB37342.1 PYGN01000075 PSN55208.1 GAKP01018211 GAKP01018206 GAKP01018203 JAC40741.1 GDHF01027793 GDHF01027140 JAI24521.1 JAI25174.1 GDHF01017424 GDHF01014498 GDHF01011975 JAI34890.1 JAI37816.1 JAI40339.1 UFQT01000821 SSX27375.1 GDHF01027990 GDHF01019235 GDHF01005964 JAI24324.1 JAI33079.1 JAI46350.1 UFQS01000410 UFQT01000410 SSX03674.1 SSX24039.1 GEDC01009839 JAS27459.1 KQ971316 EEZ98764.2 GEDC01028995 JAS08303.1 GECU01033072 GECU01012814 GECU01010734 JAS74634.1 JAS94892.1 JAS96972.1 AXCN02000015 GECZ01006241 JAS63528.1 GEBQ01005178 JAT34799.1 CH477695 EAT37153.1 CP012525 ALC43355.1 CH933809 EDW17731.1 JXUM01141253 KQ569212 KXJ68716.1 CCAG010000101 CH480817 EDW50222.1 AAAB01008888 EAA08954.3 JXJN01013627 APCN01005538 OUUW01000012 SPP87156.1 CVRI01000072 CRL07513.1 GEZM01057514 GEZM01057513 JAV72286.1 CH940647 EDW68959.2 AJWK01000428 AXCM01001019 CM000363 CM002912 EDX08970.1 KMY97174.1 AE014296 BT150200 AAF47674.1 AGS83392.1 CH902618 EDV39885.1 CH916366 EDV96689.1 DS233377 EDS29676.1 BT003289 AAO25049.1 CH954178 EDV50328.1 QCYY01002708 ROT68192.1 CH379067 EAL31337.1 CH964062 EDW78837.1 GDRN01082241 GDRN01082240 JAI61871.1 GDRN01082237 JAI61874.1 GDRN01082235 JAI61875.1 GDIQ01136489 JAL15237.1 CM000159 EDW93907.1 GDIP01123196 JAL80518.1 GFDF01008125 JAV05959.1 GFXV01003310 MBW15115.1 GDIP01108344 JAL95370.1 GFDF01008126 JAV05958.1 GDIP01099845 JAM03870.1 KK853149 KDR10650.1 GBHO01008601 GBHO01002625 GBRD01009873 GDHC01014726 GDHC01002541 JAG35003.1 JAG40979.1 JAG55951.1 JAQ03903.1 JAQ16088.1 LRGB01000781 KZS16017.1 GDIQ01009115 JAN85622.1 GDIP01093402 JAM10313.1 GDIQ01109537 JAL42189.1 GDIQ01219794 JAK31931.1 GDIP01093403 JAM10312.1 GDIQ01112888 JAL38838.1 GDIQ01230436 JAK21289.1 GDIP01194091 JAJ29311.1 GDIP01111742 JAL91972.1 GDIP01162339 JAJ61063.1 GGMR01007430 MBY20049.1 GDIP01066805 JAM36910.1 GDIP01140753 JAL62961.1 GDIP01011521 JAM92194.1 GDIP01055408 JAM48307.1 GDIP01141949 JAL61765.1 GDIP01076037 JAM27678.1 ADMH02001019 ETN64431.1 AAGJ04065343 GGMS01004613 MBY73816.1
PZC73136.1 GAIX01006679 JAA85881.1 KQ459249 KPJ02223.1 AGBW02014604 OWR41350.1 KQ461108 KPJ08964.1 GBXI01006442 JAD07850.1 ATLV01013150 KE524842 KFB37342.1 PYGN01000075 PSN55208.1 GAKP01018211 GAKP01018206 GAKP01018203 JAC40741.1 GDHF01027793 GDHF01027140 JAI24521.1 JAI25174.1 GDHF01017424 GDHF01014498 GDHF01011975 JAI34890.1 JAI37816.1 JAI40339.1 UFQT01000821 SSX27375.1 GDHF01027990 GDHF01019235 GDHF01005964 JAI24324.1 JAI33079.1 JAI46350.1 UFQS01000410 UFQT01000410 SSX03674.1 SSX24039.1 GEDC01009839 JAS27459.1 KQ971316 EEZ98764.2 GEDC01028995 JAS08303.1 GECU01033072 GECU01012814 GECU01010734 JAS74634.1 JAS94892.1 JAS96972.1 AXCN02000015 GECZ01006241 JAS63528.1 GEBQ01005178 JAT34799.1 CH477695 EAT37153.1 CP012525 ALC43355.1 CH933809 EDW17731.1 JXUM01141253 KQ569212 KXJ68716.1 CCAG010000101 CH480817 EDW50222.1 AAAB01008888 EAA08954.3 JXJN01013627 APCN01005538 OUUW01000012 SPP87156.1 CVRI01000072 CRL07513.1 GEZM01057514 GEZM01057513 JAV72286.1 CH940647 EDW68959.2 AJWK01000428 AXCM01001019 CM000363 CM002912 EDX08970.1 KMY97174.1 AE014296 BT150200 AAF47674.1 AGS83392.1 CH902618 EDV39885.1 CH916366 EDV96689.1 DS233377 EDS29676.1 BT003289 AAO25049.1 CH954178 EDV50328.1 QCYY01002708 ROT68192.1 CH379067 EAL31337.1 CH964062 EDW78837.1 GDRN01082241 GDRN01082240 JAI61871.1 GDRN01082237 JAI61874.1 GDRN01082235 JAI61875.1 GDIQ01136489 JAL15237.1 CM000159 EDW93907.1 GDIP01123196 JAL80518.1 GFDF01008125 JAV05959.1 GFXV01003310 MBW15115.1 GDIP01108344 JAL95370.1 GFDF01008126 JAV05958.1 GDIP01099845 JAM03870.1 KK853149 KDR10650.1 GBHO01008601 GBHO01002625 GBRD01009873 GDHC01014726 GDHC01002541 JAG35003.1 JAG40979.1 JAG55951.1 JAQ03903.1 JAQ16088.1 LRGB01000781 KZS16017.1 GDIQ01009115 JAN85622.1 GDIP01093402 JAM10313.1 GDIQ01109537 JAL42189.1 GDIQ01219794 JAK31931.1 GDIP01093403 JAM10312.1 GDIQ01112888 JAL38838.1 GDIQ01230436 JAK21289.1 GDIP01194091 JAJ29311.1 GDIP01111742 JAL91972.1 GDIP01162339 JAJ61063.1 GGMR01007430 MBY20049.1 GDIP01066805 JAM36910.1 GDIP01140753 JAL62961.1 GDIP01011521 JAM92194.1 GDIP01055408 JAM48307.1 GDIP01141949 JAL61765.1 GDIP01076037 JAM27678.1 ADMH02001019 ETN64431.1 AAGJ04065343 GGMS01004613 MBY73816.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000030765
+ More
UP000245037 UP000007266 UP000075886 UP000008820 UP000095301 UP000075880 UP000078200 UP000092553 UP000075900 UP000075903 UP000009192 UP000069940 UP000249989 UP000092444 UP000092445 UP000001292 UP000075882 UP000007062 UP000092460 UP000075902 UP000075840 UP000076407 UP000092443 UP000076408 UP000268350 UP000183832 UP000008792 UP000092461 UP000095300 UP000075883 UP000000304 UP000000803 UP000007801 UP000075920 UP000001070 UP000002320 UP000008711 UP000283509 UP000001819 UP000007798 UP000192221 UP000002282 UP000091820 UP000027135 UP000076858 UP000000673 UP000075884 UP000007110 UP000069272
UP000245037 UP000007266 UP000075886 UP000008820 UP000095301 UP000075880 UP000078200 UP000092553 UP000075900 UP000075903 UP000009192 UP000069940 UP000249989 UP000092444 UP000092445 UP000001292 UP000075882 UP000007062 UP000092460 UP000075902 UP000075840 UP000076407 UP000092443 UP000076408 UP000268350 UP000183832 UP000008792 UP000092461 UP000095300 UP000075883 UP000000304 UP000000803 UP000007801 UP000075920 UP000001070 UP000002320 UP000008711 UP000283509 UP000001819 UP000007798 UP000192221 UP000002282 UP000091820 UP000027135 UP000076858 UP000000673 UP000075884 UP000007110 UP000069272
Pfam
Interpro
IPR006195
aa-tRNA-synth_II
+ More
IPR002316 Pro-tRNA-ligase_IIa
IPR002314 aa-tRNA-synt_IIb
IPR036621 Anticodon-bd_dom_sf
IPR033730 ProRS_core_prok
IPR004154 Anticodon-bd
IPR032675 LRR_dom_sf
IPR003591 Leu-rich_rpt_typical-subtyp
IPR001611 Leu-rich_rpt
IPR008389 ATPase_V0-cplx_e1/e2_su
IPR000711 ATPase_OSCP/dsu
IPR026015 ATP_synth_OSCP/delta_N_sf
IPR036754 YbaK/aa-tRNA-synt-asso_dom_sf
IPR002316 Pro-tRNA-ligase_IIa
IPR002314 aa-tRNA-synt_IIb
IPR036621 Anticodon-bd_dom_sf
IPR033730 ProRS_core_prok
IPR004154 Anticodon-bd
IPR032675 LRR_dom_sf
IPR003591 Leu-rich_rpt_typical-subtyp
IPR001611 Leu-rich_rpt
IPR008389 ATPase_V0-cplx_e1/e2_su
IPR000711 ATPase_OSCP/dsu
IPR026015 ATP_synth_OSCP/delta_N_sf
IPR036754 YbaK/aa-tRNA-synt-asso_dom_sf
SUPFAM
SSF47928
SSF47928
Gene 3D
ProteinModelPortal
H9JJB1
A0A2A4JKU7
A0A2H1WFD1
A0A2W1BJX6
S4P313
A0A194Q9P0
+ More
A0A212EIR1 A0A194QUG2 A0A0A1XAV5 A0A084VH98 A0A2P8ZFE6 A0A034VBT6 A0A0K8UEV5 A0A0K8V8I2 A0A336MAS1 A0A0K8UCF6 A0A336M592 A0A1B6DP72 D6WC57 A0A1B6C484 A0A1B6HIW0 A0A182QU97 A0A1B6GM72 A0A1B6MFX8 Q0IEE6 A0A1I8MRT3 A0A182JII1 A0A1A9VHK3 A0A0M4EN92 A0A1I8JUZ4 A0A182V1E4 B4KYF2 A0A182G3B0 A0A1B0FBZ7 A0A1B0AG19 B4HTF1 A0A182KXJ1 Q7QAP8 A0A1B0BFR2 A0A182U533 A0A1I8JTP7 A0A2C9H8K8 A0A1A9Y6U0 A0A182Y7C6 A0A3B0KMU1 A0A1J1J937 A0A1Y1LL22 B4LDT6 A0A1B0C8D2 A0A1I8P2Z3 A0A182M4Y8 B4QNK7 Q9VZY9 B3M7I7 A0A1Y9IVS2 B4IZD4 B0XIT5 Q86P96 B3NBK6 A0A3R7PED5 Q29FB8 B4N398 A0A0P4W0H3 A0A0P4WKV3 A0A0P4WC38 A0A0P5P8E5 A0A1W4W297 B4PDC7 A0A1A9WFJ7 A0A0P5TR34 A0A1L8DHL5 A0A2H8TLW5 A0A0P5V1L7 A0A1L8DHM3 A0A0P5VHJ3 A0A067QQ20 A0A0A9YV78 A0A164Z717 A0A0P6I8D1 A0A0P5VYF4 A0A0P5R0S5 A0A0P5I0G0 A0A0P5WUP6 A0A0P5QKQ0 A0A0P5HAK0 A0A0P5AVD6 A0A0P5UNQ1 A0A0P5D1Q1 A0A2S2NSA3 A0A0P5XW94 A0A0P5SEW4 A0A0P6BPZ7 A0A0P5YND8 A0A0P5SQ69 A0A0P5X868 W5JJZ9 A0A182N4R5 W4XZ81 A0A182FNS6 A0A2S2Q7V4
A0A212EIR1 A0A194QUG2 A0A0A1XAV5 A0A084VH98 A0A2P8ZFE6 A0A034VBT6 A0A0K8UEV5 A0A0K8V8I2 A0A336MAS1 A0A0K8UCF6 A0A336M592 A0A1B6DP72 D6WC57 A0A1B6C484 A0A1B6HIW0 A0A182QU97 A0A1B6GM72 A0A1B6MFX8 Q0IEE6 A0A1I8MRT3 A0A182JII1 A0A1A9VHK3 A0A0M4EN92 A0A1I8JUZ4 A0A182V1E4 B4KYF2 A0A182G3B0 A0A1B0FBZ7 A0A1B0AG19 B4HTF1 A0A182KXJ1 Q7QAP8 A0A1B0BFR2 A0A182U533 A0A1I8JTP7 A0A2C9H8K8 A0A1A9Y6U0 A0A182Y7C6 A0A3B0KMU1 A0A1J1J937 A0A1Y1LL22 B4LDT6 A0A1B0C8D2 A0A1I8P2Z3 A0A182M4Y8 B4QNK7 Q9VZY9 B3M7I7 A0A1Y9IVS2 B4IZD4 B0XIT5 Q86P96 B3NBK6 A0A3R7PED5 Q29FB8 B4N398 A0A0P4W0H3 A0A0P4WKV3 A0A0P4WC38 A0A0P5P8E5 A0A1W4W297 B4PDC7 A0A1A9WFJ7 A0A0P5TR34 A0A1L8DHL5 A0A2H8TLW5 A0A0P5V1L7 A0A1L8DHM3 A0A0P5VHJ3 A0A067QQ20 A0A0A9YV78 A0A164Z717 A0A0P6I8D1 A0A0P5VYF4 A0A0P5R0S5 A0A0P5I0G0 A0A0P5WUP6 A0A0P5QKQ0 A0A0P5HAK0 A0A0P5AVD6 A0A0P5UNQ1 A0A0P5D1Q1 A0A2S2NSA3 A0A0P5XW94 A0A0P5SEW4 A0A0P6BPZ7 A0A0P5YND8 A0A0P5SQ69 A0A0P5X868 W5JJZ9 A0A182N4R5 W4XZ81 A0A182FNS6 A0A2S2Q7V4
PDB
5ZNK
E-value=9.81552e-19,
Score=223
Ontologies
GO
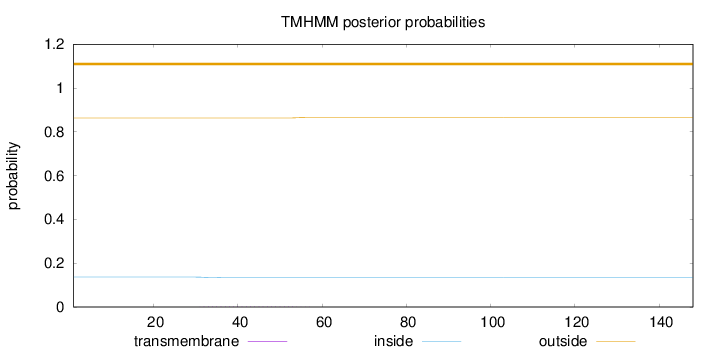
Topology
Length:
148
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0462300000000001
Exp number, first 60 AAs:
0.04603
Total prob of N-in:
0.13681
outside
1 - 148
Population Genetic Test Statistics
Pi
282.572178
Theta
187.949645
Tajima's D
1.592993
CLR
0.072007
CSRT
0.804859757012149
Interpretation
Uncertain
