Gene
KWMTBOMO14676
Pre Gene Modal
BGIBMGA009611
Annotation
PREDICTED:_protein_slit_isoform_X2_[Bombyx_mori]
Full name
Protein slit
+ More
Slit homolog 1 protein
Slit homolog 1 protein
Alternative Name
Multiple epidermal growth factor-like domains protein 4
Location in the cell
Extracellular Reliability : 2.28
Sequence
CDS
ATGAAGGTCCTACTCCTCTTGGCTCTGAGCTGGGCGACCGCCGCGGCCACCTGCCCATGGGCTTGCTCCTGCAGACCACCGGCGGCTGACTGCGCGCACAGAGGACTCCTGCATGTACCGAAGCGCTTGCCTCCGGATTCGCATAGACTAGACCTTCAGGGTAACAATATCAGCATAGTCTTTCAGAGCGACTTCCAGAACATCAAGGAGCTTAAAGTTTTACAGCTGTCGGAGAACCAGATTCACACGATTGAGAGAGACGCCTTCCTCGAGCTGACGTCACTTGAACGATTAGATCTGAGCCGAAACGAGTTGACAGCGATCAGTCGCCGCACGTTCCGAGGATTGACCGCTCTGAAGAGTCTACACCTCGACGGGAACCAGCTGAAATGCATCGACGAGAAAGCTCTGGAGAACTTGAAAAGCCTGGAAGTTCTGACTTTAAATAACAACAACCTAACGTATTTGTCGTTGGAGCCGGTCACCGTCTCGCGACTGAGCACGCTGCGGCTGAGCGACAATCCGGTTGTCTGCGACTGTCGCGTCGCAAGACTGTCCACAGCTGTCCGGTCTGCTGGGATCATGGGCGTCGGCGCGAGGTGTCAAGCTCCTGCAGCTCTACGTGGCTCGTTGCTGACCGACTTAGAGCCTCACGAACTCATCTGCAGTGGACCTACAACAGCTTCGTCTGGTGAATGCTCAGCGGAGCCCCGCTGCCCGCCACCCTGTCGCTGCACTCCTGAGGGTATAGTGGACTGCAGGGAGAAGATGCTCTCAGAGCTGCCACCTACTATACCTCACAGAGCTACGGAGATTCGCTTGGAACAGAATGAGATAACCGAAGTCGGCAGCGGCGCCTTCTCATCCATAAAGAGGGTGACCAGGATCGACCTGTCCAACAACAAGATCGCCAAAATCTCAGCGGACACATTCCAAGGACTCGTACATTTAACCTCACTAGTCTTGTACGGAAATAAAATAAAGGATTTGCCGTCGGGAGTATTCCACGGGCTGGGATCGCTCCAATTATTACTGCTGAACTCGAATGAGATTACGTGCGTCAAAAAGGACACGTTCCGTGATTTAGAGAATCTAAAATTACTGTCTTTATACGACAACAATATACGATCACTGCCGAATGGTACCTTCGATGCTTTGACGGGAATCCAGACCTTGCATTTAGGTCGGAATCCGTTCGTATGCGACTGTTCCCTCCGCTGGCTGGCTGCCTACTTGCGCAAGAATCCCATCGAGACGTCTGGTGCCAAGTGTGACTCTCCGAAGCGCATGAACAAAAAGCGAATCGACGCTCTCAGGGAAGAGAACTTCAAATGCAAGCGTGAGTTGCCATCGTAG
Protein
MKVLLLLALSWATAAATCPWACSCRPPAADCAHRGLLHVPKRLPPDSHRLDLQGNNISIVFQSDFQNIKELKVLQLSENQIHTIERDAFLELTSLERLDLSRNELTAISRRTFRGLTALKSLHLDGNQLKCIDEKALENLKSLEVLTLNNNNLTYLSLEPVTVSRLSTLRLSDNPVVCDCRVARLSTAVRSAGIMGVGARCQAPAALRGSLLTDLEPHELICSGPTTASSGECSAEPRCPPPCRCTPEGIVDCREKMLSELPPTIPHRATEIRLEQNEITEVGSGAFSSIKRVTRIDLSNNKIAKISADTFQGLVHLTSLVLYGNKIKDLPSGVFHGLGSLQLLLLNSNEITCVKKDTFRDLENLKLLSLYDNNIRSLPNGTFDALTGIQTLHLGRNPFVCDCSLRWLAAYLRKNPIETSGAKCDSPKRMNKKRIDALREENFKCKRELPS
Summary
Description
A short-range repellent, controlling axon crossing of the midline and a long-range chemorepellent, controlling mesoderm migration and patterning away from the midline. May interact with extracellular matrix molecules. Repulsive ligand for the guidance receptor roundabout (robo) and prevents inappropriate midline crossing by Robo-expressing axons.
Thought to act as molecular guidance cue in cellular migration, and function appears to be mediated by interaction with roundabout homolog receptors. During neural development involved in axonal navigation at the ventral midline of the neural tube and projection of axons to different regions. SLIT1 and SLIT2 together seem to be essential for midline guidance in the forebrain by acting as repulsive signal preventing inappropriate midline crossing by axons projecting from the olfactory bulb (By similarity).
Thought to act as molecular guidance cue in cellular migration, and function appears to be mediated by interaction with roundabout homolog receptors. During neural development involved in axonal navigation at the ventral midline of the neural tube and projection of axons to different regions (By similarity). SLIT1 and SLIT2 together seem to be essential for midline guidance in the forebrain by acting as repulsive signal preventing inappropriate midline crossing by axons projecting from the olfactory bulb.
Thought to act as molecular guidance cue in cellular migration, and function appears to be mediated by interaction with roundabout homolog receptors. During neural development involved in axonal navigation at the ventral midline of the neural tube and projection of axons to different regions. SLIT1 and SLIT2 together seem to be essential for midline guidance in the forebrain by acting as repulsive signal preventing inappropriate midline crossing by axons projecting from the olfactory bulb (By similarity).
Thought to act as molecular guidance cue in cellular migration, and function appears to be mediated by interaction with roundabout homolog receptors. During neural development involved in axonal navigation at the ventral midline of the neural tube and projection of axons to different regions (By similarity). SLIT1 and SLIT2 together seem to be essential for midline guidance in the forebrain by acting as repulsive signal preventing inappropriate midline crossing by axons projecting from the olfactory bulb.
Subunit
Interacts with robo.
Interacts with ROBO1 (By similarity) and GREM1.
Interacts with ROBO1 and GREM1.
Interacts with ROBO1 (By similarity) and GREM1.
Interacts with ROBO1 and GREM1.
Keywords
3D-structure
Alternative splicing
Complete proteome
Developmental protein
Differentiation
Disulfide bond
EGF-like domain
Glycoprotein
Leucine-rich repeat
Neurogenesis
Reference proteome
Repeat
Secreted
Signal
Polymorphism
Feature
chain Protein slit
splice variant In isoform A and isoform B.
sequence variant In dbSNP:rs2817673.
splice variant In isoform A and isoform B.
sequence variant In dbSNP:rs2817673.
Uniprot
H9JJB0
A0A2H1V322
A0A2W1BK68
A0A194QFI1
A0A034V431
A0A1B6KZT4
+ More
A0A1B6IAM4 A0A1W5LU06 W8AUW0 A0A1B6MA00 A0A1Y1MIY6 A0A1Y1MHN4 A0A034V6T1 W8BHF3 W8B2B3 A0A034V560 A0A034V1X2 E1NZD5 P24014-3 P24014-2 A0A139WM17 A0A0K8TXL8 E0VSI9 A0A0A1X6C6 A0A0A1X7M8 E3CTP8 F0JAL2 P24014 B4QGZ8 B4HSG3 A0A3B0IZ80 A0A0K8UZ73 A0A0M4EA23 A0A0K8UWZ3 A0A1I8NNR6 A0A0L0CU11 B4P6U8 E1NZD6 N6WCZ8 B3MC19 B4J4Q8 A0A0B4KES0 B3NQ10 A0A0J9REU2 A0A182Q2Z8 A0A1I8MJQ3 B4KM13 A0A1W4UZ69 B0WB86 B4LQ27 B4MNM8 A0A182N797 Q16WM1 A0A2H8TPR9 Q7QCT2 A0A1J1HFQ7 A0A232FEP7 K7INA9 A0A0P5QAF6 A0A0N8BJW4 A0A0P5QV25 A0A0P5LP38 A0A182F736 A0A0P5G8Z9 A0A0P5HSP1 A0A0P6GCF6 A0A0P5HM86 A0A0D6A818 A0A0P5ELL6 A0A0P6HUB3 A0A0P5WQ00 A0A0P6CY20 A0A0P5U8J6 A0A0P5V3A4 A0A0P5BKL2 A0A0P4YJ18 A0A0P5UWX4 A0A0P5B090 A0A0P5Z6T7 E9GML1 A0A0P5WVB3 A6YB93 A0A0P6BYS5 A0A0P5VTL1 X1WIF9 A0A2S2R2P7 A0A2J8PDR7 A0A2I2MJP8 A0A1A9YJM8 O88279-4 A0A0P5RH75 A0A182YR84 A0A182XHS7 A0A088AS83 Q5T0V4 A0A2J8PDS4 O75093-2 A0A146WBZ8 A0A147BKB3 A1A5F4
A0A1B6IAM4 A0A1W5LU06 W8AUW0 A0A1B6MA00 A0A1Y1MIY6 A0A1Y1MHN4 A0A034V6T1 W8BHF3 W8B2B3 A0A034V560 A0A034V1X2 E1NZD5 P24014-3 P24014-2 A0A139WM17 A0A0K8TXL8 E0VSI9 A0A0A1X6C6 A0A0A1X7M8 E3CTP8 F0JAL2 P24014 B4QGZ8 B4HSG3 A0A3B0IZ80 A0A0K8UZ73 A0A0M4EA23 A0A0K8UWZ3 A0A1I8NNR6 A0A0L0CU11 B4P6U8 E1NZD6 N6WCZ8 B3MC19 B4J4Q8 A0A0B4KES0 B3NQ10 A0A0J9REU2 A0A182Q2Z8 A0A1I8MJQ3 B4KM13 A0A1W4UZ69 B0WB86 B4LQ27 B4MNM8 A0A182N797 Q16WM1 A0A2H8TPR9 Q7QCT2 A0A1J1HFQ7 A0A232FEP7 K7INA9 A0A0P5QAF6 A0A0N8BJW4 A0A0P5QV25 A0A0P5LP38 A0A182F736 A0A0P5G8Z9 A0A0P5HSP1 A0A0P6GCF6 A0A0P5HM86 A0A0D6A818 A0A0P5ELL6 A0A0P6HUB3 A0A0P5WQ00 A0A0P6CY20 A0A0P5U8J6 A0A0P5V3A4 A0A0P5BKL2 A0A0P4YJ18 A0A0P5UWX4 A0A0P5B090 A0A0P5Z6T7 E9GML1 A0A0P5WVB3 A6YB93 A0A0P6BYS5 A0A0P5VTL1 X1WIF9 A0A2S2R2P7 A0A2J8PDR7 A0A2I2MJP8 A0A1A9YJM8 O88279-4 A0A0P5RH75 A0A182YR84 A0A182XHS7 A0A088AS83 Q5T0V4 A0A2J8PDS4 O75093-2 A0A146WBZ8 A0A147BKB3 A1A5F4
Pubmed
19121390
28756777
26354079
25348373
24495485
28004739
+ More
2176636 10102267 10731132 12537572 3144436 10102268 15496984 18362917 19820115 20566863 25830018 17994087 26108605 17550304 15632085 12537568 12537573 12537574 16110336 17569856 17569867 22936249 25315136 17510324 12364791 28648823 20075255 21292972 17448990 9693030 9813312 10864956 11754167 15528323 25244985 15164054 12168954 15489334 12141424 12200164 29652888 20431018
2176636 10102267 10731132 12537572 3144436 10102268 15496984 18362917 19820115 20566863 25830018 17994087 26108605 17550304 15632085 12537568 12537573 12537574 16110336 17569856 17569867 22936249 25315136 17510324 12364791 28648823 20075255 21292972 17448990 9693030 9813312 10864956 11754167 15528323 25244985 15164054 12168954 15489334 12141424 12200164 29652888 20431018
EMBL
BABH01036403
BABH01036404
ODYU01000445
SOQ35227.1
KZ150125
PZC73120.1
+ More
KQ459249 KPJ02226.1 GAKP01021703 JAC37249.1 GEBQ01023010 JAT16967.1 GECU01023737 JAS83969.1 KX034172 ANO54002.1 GAMC01013890 JAB92665.1 GEBQ01007303 JAT32674.1 GEZM01034259 JAV83846.1 GEZM01034260 JAV83845.1 GAKP01021709 JAC37243.1 GAMC01013889 JAB92666.1 GAMC01013888 JAB92667.1 GAKP01021700 JAC37252.1 GAKP01021711 JAC37241.1 BT125684 ADO14992.1 X53959 AF126540 AE013599 M23543 KQ971316 KYB28990.1 GDHF01033283 GDHF01030424 GDHF01002259 JAI19031.1 JAI21890.1 JAI50055.1 DS235751 EEB16345.1 GBXI01007851 JAD06441.1 GBXI01007604 JAD06688.1 BT125714 ADO51076.1 BT126054 ADY17753.1 CM000362 EDX07261.1 CH480816 EDW48041.1 OUUW01000001 SPP73287.1 GDHF01021411 GDHF01020476 GDHF01008854 GDHF01000960 JAI30903.1 JAI31838.1 JAI43460.1 JAI51354.1 CP012524 ALC41858.1 GDHF01032461 GDHF01027435 GDHF01021208 GDHF01019939 GDHF01001960 JAI19853.1 JAI24879.1 JAI31106.1 JAI32375.1 JAI50354.1 JRES01000018 KNC34864.1 CM000158 EDW92025.2 BT125685 ADO16251.1 CM000071 ENO01959.1 CH902619 EDV37206.2 CH916367 EDW00604.1 AGB93549.1 CH954179 EDV55857.2 CM002911 KMY94099.1 AXCN02001179 CH933808 EDW10802.2 DS231877 EDS42151.1 CH940648 EDW60350.2 CH963848 EDW73717.2 CH477561 EAT38989.1 GFXV01004351 MBW16156.1 AAAB01008859 EAA07783.5 CVRI01000002 CRK86807.1 NNAY01000318 OXU29264.1 GDIQ01117370 JAL34356.1 GDIQ01170547 JAK81178.1 GDIQ01116073 JAL35653.1 GDIQ01170548 JAK81177.1 GDIQ01251925 JAJ99799.1 GDIQ01223277 JAK28448.1 GDIQ01035346 JAN59391.1 GDIQ01231381 JAK20344.1 AB981662 BAQ58829.1 GDIP01157966 JAJ65436.1 GDIQ01029750 JAN64987.1 GDIP01083232 JAM20483.1 GDIQ01085977 GDIQ01009364 JAN08760.1 GDIP01120321 JAL83393.1 GDIP01107667 JAL96047.1 GDIP01198213 JAJ25189.1 GDIP01228066 JAI95335.1 GDIP01107668 JAL96046.1 GDIP01191860 JAJ31542.1 GDIP01050330 JAM53385.1 GL732553 EFX79275.1 GDIP01081276 JAM22439.1 EF384216 ABO93215.1 GDIP01008518 JAM95197.1 GDIP01095530 JAM08185.1 ABLF02034397 ABLF02034398 ABLF02034400 GGMS01015063 MBY84266.1 NBAG03000216 PNI82166.1 LT968152 SOU94110.1 AB011530 AF133730 AB073213 AB073214 AB073215 AB017170 GDIQ01120016 JAL31710.1 AL442123 AL512424 PNI82169.1 AB017167 AB011537 CH471066 BC146851 AY029183 GCES01059767 JAR26556.1 GEGO01004177 JAR91227.1 AAMC01095566 AAMC01095567 AAMC01095568 AAMC01095569 AAMC01095570 AAMC01095571 AAMC01095572 AAMC01095573 AAMC01095574 AAMC01095575 BC128626 BC135997 AAI28627.1
KQ459249 KPJ02226.1 GAKP01021703 JAC37249.1 GEBQ01023010 JAT16967.1 GECU01023737 JAS83969.1 KX034172 ANO54002.1 GAMC01013890 JAB92665.1 GEBQ01007303 JAT32674.1 GEZM01034259 JAV83846.1 GEZM01034260 JAV83845.1 GAKP01021709 JAC37243.1 GAMC01013889 JAB92666.1 GAMC01013888 JAB92667.1 GAKP01021700 JAC37252.1 GAKP01021711 JAC37241.1 BT125684 ADO14992.1 X53959 AF126540 AE013599 M23543 KQ971316 KYB28990.1 GDHF01033283 GDHF01030424 GDHF01002259 JAI19031.1 JAI21890.1 JAI50055.1 DS235751 EEB16345.1 GBXI01007851 JAD06441.1 GBXI01007604 JAD06688.1 BT125714 ADO51076.1 BT126054 ADY17753.1 CM000362 EDX07261.1 CH480816 EDW48041.1 OUUW01000001 SPP73287.1 GDHF01021411 GDHF01020476 GDHF01008854 GDHF01000960 JAI30903.1 JAI31838.1 JAI43460.1 JAI51354.1 CP012524 ALC41858.1 GDHF01032461 GDHF01027435 GDHF01021208 GDHF01019939 GDHF01001960 JAI19853.1 JAI24879.1 JAI31106.1 JAI32375.1 JAI50354.1 JRES01000018 KNC34864.1 CM000158 EDW92025.2 BT125685 ADO16251.1 CM000071 ENO01959.1 CH902619 EDV37206.2 CH916367 EDW00604.1 AGB93549.1 CH954179 EDV55857.2 CM002911 KMY94099.1 AXCN02001179 CH933808 EDW10802.2 DS231877 EDS42151.1 CH940648 EDW60350.2 CH963848 EDW73717.2 CH477561 EAT38989.1 GFXV01004351 MBW16156.1 AAAB01008859 EAA07783.5 CVRI01000002 CRK86807.1 NNAY01000318 OXU29264.1 GDIQ01117370 JAL34356.1 GDIQ01170547 JAK81178.1 GDIQ01116073 JAL35653.1 GDIQ01170548 JAK81177.1 GDIQ01251925 JAJ99799.1 GDIQ01223277 JAK28448.1 GDIQ01035346 JAN59391.1 GDIQ01231381 JAK20344.1 AB981662 BAQ58829.1 GDIP01157966 JAJ65436.1 GDIQ01029750 JAN64987.1 GDIP01083232 JAM20483.1 GDIQ01085977 GDIQ01009364 JAN08760.1 GDIP01120321 JAL83393.1 GDIP01107667 JAL96047.1 GDIP01198213 JAJ25189.1 GDIP01228066 JAI95335.1 GDIP01107668 JAL96046.1 GDIP01191860 JAJ31542.1 GDIP01050330 JAM53385.1 GL732553 EFX79275.1 GDIP01081276 JAM22439.1 EF384216 ABO93215.1 GDIP01008518 JAM95197.1 GDIP01095530 JAM08185.1 ABLF02034397 ABLF02034398 ABLF02034400 GGMS01015063 MBY84266.1 NBAG03000216 PNI82166.1 LT968152 SOU94110.1 AB011530 AF133730 AB073213 AB073214 AB073215 AB017170 GDIQ01120016 JAL31710.1 AL442123 AL512424 PNI82169.1 AB017167 AB011537 CH471066 BC146851 AY029183 GCES01059767 JAR26556.1 GEGO01004177 JAR91227.1 AAMC01095566 AAMC01095567 AAMC01095568 AAMC01095569 AAMC01095570 AAMC01095571 AAMC01095572 AAMC01095573 AAMC01095574 AAMC01095575 BC128626 BC135997 AAI28627.1
Proteomes
UP000005204
UP000053268
UP000000803
UP000007266
UP000009046
UP000000304
+ More
UP000001292 UP000268350 UP000092553 UP000095300 UP000037069 UP000002282 UP000001819 UP000007801 UP000001070 UP000008711 UP000075886 UP000095301 UP000009192 UP000192221 UP000002320 UP000008792 UP000007798 UP000075884 UP000008820 UP000007062 UP000183832 UP000215335 UP000002358 UP000069272 UP000000305 UP000007819 UP000092443 UP000002494 UP000076408 UP000076407 UP000005203 UP000005640 UP000008143
UP000001292 UP000268350 UP000092553 UP000095300 UP000037069 UP000002282 UP000001819 UP000007801 UP000001070 UP000008711 UP000075886 UP000095301 UP000009192 UP000192221 UP000002320 UP000008792 UP000007798 UP000075884 UP000008820 UP000007062 UP000183832 UP000215335 UP000002358 UP000069272 UP000000305 UP000007819 UP000092443 UP000002494 UP000076408 UP000076407 UP000005203 UP000005640 UP000008143
Pfam
Interpro
IPR000372
LRRNT
+ More
IPR000742 EGF-like_dom
IPR001791 Laminin_G
IPR013032 EGF-like_CS
IPR018097 EGF_Ca-bd_CS
IPR003591 Leu-rich_rpt_typical-subtyp
IPR001611 Leu-rich_rpt
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR013320 ConA-like_dom_sf
IPR001881 EGF-like_Ca-bd_dom
IPR032675 LRR_dom_sf
IPR000483 Cys-rich_flank_reg_C
IPR006207 Cys_knot_C
IPR026906 LRR_5
IPR009030 Growth_fac_rcpt_cys_sf
IPR036047 F-box-like_dom_sf
IPR003645 Fol_N
IPR000742 EGF-like_dom
IPR001791 Laminin_G
IPR013032 EGF-like_CS
IPR018097 EGF_Ca-bd_CS
IPR003591 Leu-rich_rpt_typical-subtyp
IPR001611 Leu-rich_rpt
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR013320 ConA-like_dom_sf
IPR001881 EGF-like_Ca-bd_dom
IPR032675 LRR_dom_sf
IPR000483 Cys-rich_flank_reg_C
IPR006207 Cys_knot_C
IPR026906 LRR_5
IPR009030 Growth_fac_rcpt_cys_sf
IPR036047 F-box-like_dom_sf
IPR003645 Fol_N
Gene 3D
ProteinModelPortal
H9JJB0
A0A2H1V322
A0A2W1BK68
A0A194QFI1
A0A034V431
A0A1B6KZT4
+ More
A0A1B6IAM4 A0A1W5LU06 W8AUW0 A0A1B6MA00 A0A1Y1MIY6 A0A1Y1MHN4 A0A034V6T1 W8BHF3 W8B2B3 A0A034V560 A0A034V1X2 E1NZD5 P24014-3 P24014-2 A0A139WM17 A0A0K8TXL8 E0VSI9 A0A0A1X6C6 A0A0A1X7M8 E3CTP8 F0JAL2 P24014 B4QGZ8 B4HSG3 A0A3B0IZ80 A0A0K8UZ73 A0A0M4EA23 A0A0K8UWZ3 A0A1I8NNR6 A0A0L0CU11 B4P6U8 E1NZD6 N6WCZ8 B3MC19 B4J4Q8 A0A0B4KES0 B3NQ10 A0A0J9REU2 A0A182Q2Z8 A0A1I8MJQ3 B4KM13 A0A1W4UZ69 B0WB86 B4LQ27 B4MNM8 A0A182N797 Q16WM1 A0A2H8TPR9 Q7QCT2 A0A1J1HFQ7 A0A232FEP7 K7INA9 A0A0P5QAF6 A0A0N8BJW4 A0A0P5QV25 A0A0P5LP38 A0A182F736 A0A0P5G8Z9 A0A0P5HSP1 A0A0P6GCF6 A0A0P5HM86 A0A0D6A818 A0A0P5ELL6 A0A0P6HUB3 A0A0P5WQ00 A0A0P6CY20 A0A0P5U8J6 A0A0P5V3A4 A0A0P5BKL2 A0A0P4YJ18 A0A0P5UWX4 A0A0P5B090 A0A0P5Z6T7 E9GML1 A0A0P5WVB3 A6YB93 A0A0P6BYS5 A0A0P5VTL1 X1WIF9 A0A2S2R2P7 A0A2J8PDR7 A0A2I2MJP8 A0A1A9YJM8 O88279-4 A0A0P5RH75 A0A182YR84 A0A182XHS7 A0A088AS83 Q5T0V4 A0A2J8PDS4 O75093-2 A0A146WBZ8 A0A147BKB3 A1A5F4
A0A1B6IAM4 A0A1W5LU06 W8AUW0 A0A1B6MA00 A0A1Y1MIY6 A0A1Y1MHN4 A0A034V6T1 W8BHF3 W8B2B3 A0A034V560 A0A034V1X2 E1NZD5 P24014-3 P24014-2 A0A139WM17 A0A0K8TXL8 E0VSI9 A0A0A1X6C6 A0A0A1X7M8 E3CTP8 F0JAL2 P24014 B4QGZ8 B4HSG3 A0A3B0IZ80 A0A0K8UZ73 A0A0M4EA23 A0A0K8UWZ3 A0A1I8NNR6 A0A0L0CU11 B4P6U8 E1NZD6 N6WCZ8 B3MC19 B4J4Q8 A0A0B4KES0 B3NQ10 A0A0J9REU2 A0A182Q2Z8 A0A1I8MJQ3 B4KM13 A0A1W4UZ69 B0WB86 B4LQ27 B4MNM8 A0A182N797 Q16WM1 A0A2H8TPR9 Q7QCT2 A0A1J1HFQ7 A0A232FEP7 K7INA9 A0A0P5QAF6 A0A0N8BJW4 A0A0P5QV25 A0A0P5LP38 A0A182F736 A0A0P5G8Z9 A0A0P5HSP1 A0A0P6GCF6 A0A0P5HM86 A0A0D6A818 A0A0P5ELL6 A0A0P6HUB3 A0A0P5WQ00 A0A0P6CY20 A0A0P5U8J6 A0A0P5V3A4 A0A0P5BKL2 A0A0P4YJ18 A0A0P5UWX4 A0A0P5B090 A0A0P5Z6T7 E9GML1 A0A0P5WVB3 A6YB93 A0A0P6BYS5 A0A0P5VTL1 X1WIF9 A0A2S2R2P7 A0A2J8PDR7 A0A2I2MJP8 A0A1A9YJM8 O88279-4 A0A0P5RH75 A0A182YR84 A0A182XHS7 A0A088AS83 Q5T0V4 A0A2J8PDS4 O75093-2 A0A146WBZ8 A0A147BKB3 A1A5F4
PDB
2V9T
E-value=9.94909e-56,
Score=549
Ontologies
GO
GO:0005509
GO:0016021
GO:0005576
GO:0048495
GO:0007502
GO:0008078
GO:0035050
GO:0005886
GO:0007427
GO:0030335
GO:0007509
GO:0022409
GO:0003151
GO:0016201
GO:0007411
GO:0050929
GO:0007432
GO:0030182
GO:0071666
GO:0008347
GO:0035385
GO:0001764
GO:0016199
GO:2000274
GO:0008201
GO:0008406
GO:0010632
GO:0048813
GO:0004714
GO:0007420
GO:0050919
GO:0051964
GO:0043395
GO:0048843
GO:0021510
GO:0005615
GO:0033563
GO:0008045
GO:0022028
GO:0048846
GO:0048853
GO:0031290
GO:0007097
GO:0005623
GO:0045773
GO:0021602
GO:0005515
GO:0003677
GO:0004677
GO:0005524
GO:0005634
GO:0006303
GO:0003824
GO:0008152
GO:0016491
GO:0015930
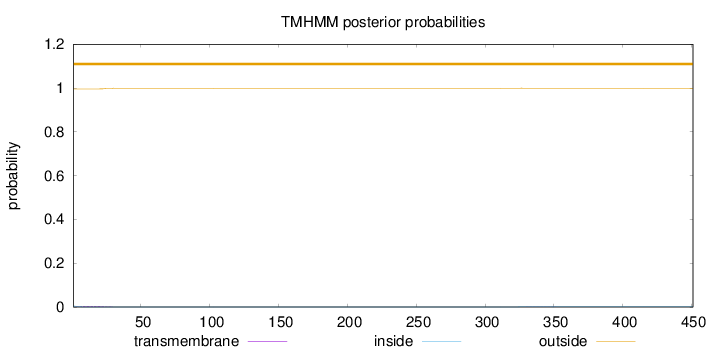
Topology
Subcellular location
Secreted
SignalP
Position: 1 - 16,
Likelihood: 0.997945
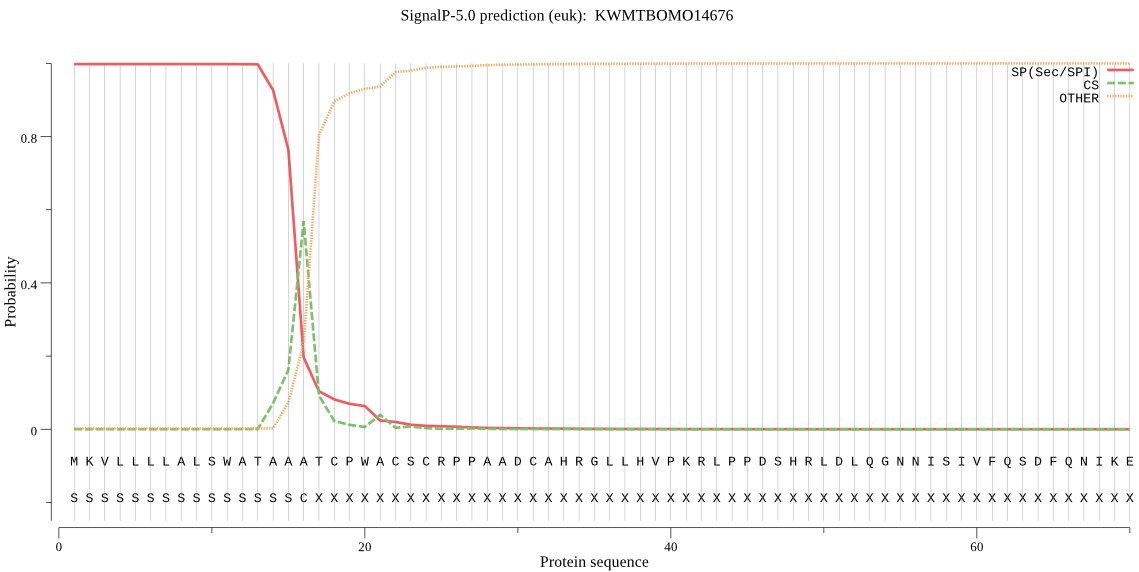
Length:
451
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0915199999999999
Exp number, first 60 AAs:
0.08155
Total prob of N-in:
0.00411
outside
1 - 451
Population Genetic Test Statistics
Pi
244.265883
Theta
156.643071
Tajima's D
1.672801
CLR
0.075635
CSRT
0.822308884555772
Interpretation
Uncertain
