Gene
KWMTBOMO14671 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009610
Annotation
serine_protease_HP21_precursor_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 4.344
Sequence
CDS
ATGGATCAATCAATATATTTGTTGTTAATGCTAGTGTTATTTGTGTGTGTTCATTGCGAATTCGAGGGTGAAGAATGTAAAAAAGGTAATTTATTAGGAGTCTGTACGAATATAAGGAAATGTCAGTCCGCGCTGAACGACATCAGAAACAGGAAATCACCACAGATTTGTTCGTTCGACAACGCGGATCCTGTTGTGTGCTGCTTTGATAACTCGATCAGTTCACGGGCTCCCCTCGCCACAACAACGCGTCGTACGCCGAGTTCTACCACAACGGAGTACGTACCACCCTCCTATGATTACCAGTCCAACAATGGCGACAAGAAATGCGAGGATGTACCGGCGGATCTGACGTCTCCCAAGACCGGGCAGAAGGCGTGGGACAAATGTATTGAATATCAAGAGCAGCTCGTGTATCCGTGTGAAAAGGGCGTCGCGCTCACCGGGGAGATCAGCCGCAGTAAGCACTGCCACCACGACGCGGACGAGCTCATCATTGGCGGCACCGATGCAGGCGTCAACGAATATCCGCACATGGTCCTGCTCGGCTACGGTGACGATGTCGCGAACATCCAGTGGCTGTGCGGGGGGGTGCTCATCAGTGAGCGCTTCGTGCTCACAGCCGGACACTGCCTATCCAGTAGAGAGGTTGGCGCAGTCAGATACGTGTATATCGGAGCCCTCGCCCGGCACGAGACCACGAACCCGTCCCGCCGCTACGCAGTCATCCGCGCTCACAGACACCCCGACTACAAGCCGCCTAGCAAGTATAACGACATCGCGCTGTTAGAGCTAGATCGACAGGTCCCGCTGGACCAGTACACAGTCCCGGCGTGTCTGCACACGGGAGACGCCGTCAATGACGAGCGGGCCTCCGCCACCGGCTGGGGACTCACCGAGAACCGGGGCTCCACGTCGGACGTGCTGCAGAAGGTGGTACTAACTAAGTTCACGTCGACGGAATGCTCCGAGAAGTACCCGACCAATCGCAACATGAAGCGCGGGTTCGACCCTCGGACTCAGATGTGCTACGGAGACAGGACATTGTCCAGAGACACTTGTCAGGGCGATAGCGGAGGTCCTATACAGATAAAAAGCAAGAAGATGGACTGTATGTACGTCGTGATAGGCGTGACGTCGTTTGGTCGGGCCTGTGGTTACGCGGGCGAGCCCGGGATCTACACCAGGGTCAGCCACTACGTACCCTGGATAGAGAGTGTCGTCTGGCCTTAG
Protein
MDQSIYLLLMLVLFVCVHCEFEGEECKKGNLLGVCTNIRKCQSALNDIRNRKSPQICSFDNADPVVCCFDNSISSRAPLATTTRRTPSSTTTEYVPPSYDYQSNNGDKKCEDVPADLTSPKTGQKAWDKCIEYQEQLVYPCEKGVALTGEISRSKHCHHDADELIIGGTDAGVNEYPHMVLLGYGDDVANIQWLCGGVLISERFVLTAGHCLSSREVGAVRYVYIGALARHETTNPSRRYAVIRAHRHPDYKPPSKYNDIALLELDRQVPLDQYTVPACLHTGDAVNDERASATGWGLTENRGSTSDVLQKVVLTKFTSTECSEKYPTNRNMKRGFDPRTQMCYGDRTLSRDTCQGDSGGPIQIKSKKMDCMYVVIGVTSFGRACGYAGEPGIYTRVSHYVPWIESVVWP
Summary
Similarity
Belongs to the peptidase S1 family.
Belongs to the PI3/PI4-kinase family.
Belongs to the PI3/PI4-kinase family.
Uniprot
F5BZV1
H9JJA9
Q5MPB3
A0A0N1PHF7
S4NXP2
A0A2A4K236
+ More
A0A2H1VS51 Q5MPB2 A0A3S2NPZ4 A0A194Q9Z8 O44331 A0A194QB71 A0A346RAE0 Q5MPC1 A0A194Q9Q9 A0A212F6Q1 A0A0N0PDL6 A0A2W1B2N2 A0A2W1B0L2 A0A2H1VJ99 A0A2A4K2L1 A0A2W1B9D9 A0A0N1IHL2 A0A2A4K714 A0A2H1VFF0 Q5MPB6 A0A2W1BFG9 A0A212F107 A0A2W1BHZ8 A0A2W1AYZ6 A0A2A4JLI0 A0A2A4JMC3 A0A2H1VJG7 A0A2H1V6L6 A0A2A4J7N6 A0A2H1W424 A0A2W1B9E2 U5ESE3 A0A2W1BD72 H9JLA5 A0A2A4IWJ7 A0A2A4J4P7 A0A2H1VSY3 A0A2H1VTL8 B0W1A9 A0A1Q3FNX1 A0A2W1BGC1 A0A2A4KAM6 Q16NE9 A0A2J7Q4L6 A0A336LYA5 D6WCU1 A0A2H1VRC0 A0A182XUQ8 A0A1Y1KAU3 A0A182IIN2 A0A182VNZ6 Q5TNC5 A0A182THH3 A0A1Y1KPC1 B1B5K0 A0A182KNG1 A0A182Y982 A0A1B6GBU6 A0A2W1BCP3 A0A224XHV2 A0A1B6LG03 A0A067RHT4 A0A1B6L129 A0A182S507 A0A084VYF5 A0A182JES5 A0A1B6LJF7 A0A182NZ25 A0A2A4J0N6 A0A1B6DI71 A0A182FZY7 A0A0U4B511 W5JRY9 A0A1L8DQW0 A0A182WR24 A0A182Q673 A0A2W1B5J0 A0A1B0CS99 A0A182MQJ3 V5GMH2 A0A2H8TED5 A0A2H8TTU6
A0A2H1VS51 Q5MPB2 A0A3S2NPZ4 A0A194Q9Z8 O44331 A0A194QB71 A0A346RAE0 Q5MPC1 A0A194Q9Q9 A0A212F6Q1 A0A0N0PDL6 A0A2W1B2N2 A0A2W1B0L2 A0A2H1VJ99 A0A2A4K2L1 A0A2W1B9D9 A0A0N1IHL2 A0A2A4K714 A0A2H1VFF0 Q5MPB6 A0A2W1BFG9 A0A212F107 A0A2W1BHZ8 A0A2W1AYZ6 A0A2A4JLI0 A0A2A4JMC3 A0A2H1VJG7 A0A2H1V6L6 A0A2A4J7N6 A0A2H1W424 A0A2W1B9E2 U5ESE3 A0A2W1BD72 H9JLA5 A0A2A4IWJ7 A0A2A4J4P7 A0A2H1VSY3 A0A2H1VTL8 B0W1A9 A0A1Q3FNX1 A0A2W1BGC1 A0A2A4KAM6 Q16NE9 A0A2J7Q4L6 A0A336LYA5 D6WCU1 A0A2H1VRC0 A0A182XUQ8 A0A1Y1KAU3 A0A182IIN2 A0A182VNZ6 Q5TNC5 A0A182THH3 A0A1Y1KPC1 B1B5K0 A0A182KNG1 A0A182Y982 A0A1B6GBU6 A0A2W1BCP3 A0A224XHV2 A0A1B6LG03 A0A067RHT4 A0A1B6L129 A0A182S507 A0A084VYF5 A0A182JES5 A0A1B6LJF7 A0A182NZ25 A0A2A4J0N6 A0A1B6DI71 A0A182FZY7 A0A0U4B511 W5JRY9 A0A1L8DQW0 A0A182WR24 A0A182Q673 A0A2W1B5J0 A0A1B0CS99 A0A182MQJ3 V5GMH2 A0A2H8TED5 A0A2H8TTU6
Pubmed
EMBL
JF431073
AEB21638.1
BABH01036383
BABH01036384
BABH01036385
BABH01036386
+ More
BABH01036387 BABH01036388 BABH01036389 BABH01036390 BABH01036391 AY672797 AAV91019.1 KQ460127 KPJ17583.1 GAIX01012077 JAA80483.1 NWSH01000257 PCG77964.1 ODYU01004095 SOQ43606.1 AY672798 AAV91020.1 RSAL01000491 RVE41548.1 KQ459249 KPJ02244.1 AF017664 AAB94558.1 KPJ02240.1 MH200930 AXS59140.1 AY672789 AAV91011.1 KPJ02243.1 AGBW02009991 OWR49389.1 KQ460130 KPJ17530.1 KZ150408 PZC70992.1 KZ150503 PZC70659.1 ODYU01002875 SOQ40903.1 NWSH01000265 PCG77882.1 KZ150484 PZC70714.1 KPJ17576.1 NWSH01000060 PCG80065.1 ODYU01002282 SOQ39557.1 AY672794 AAV91016.1 KZ150189 PZC72424.1 AGBW02010978 OWR47436.1 KZ150344 PZC71293.1 KZ150652 PZC70412.1 NWSH01001045 PCG72935.1 PCG72936.1 ODYU01002903 SOQ40975.1 ODYU01000940 SOQ36457.1 NWSH01002560 PCG67981.1 ODYU01006166 SOQ47783.1 KZ150324 PZC71401.1 GANO01003282 JAB56589.1 PZC71287.1 BABH01038780 NWSH01005248 PCG64337.1 NWSH01003268 PCG66648.1 ODYU01004086 SOQ43592.1 SOQ43594.1 DS231821 EDS44979.1 GFDL01005857 JAV29188.1 KZ150249 PZC71840.1 NWSH01000008 PCG80964.1 CH477825 EAT35883.2 NEVH01018378 PNF23527.1 UFQT01000075 UFQT01000299 SSX19399.1 SSX22964.1 KQ971311 EEZ99357.2 ODYU01003947 SOQ43326.1 GEZM01092351 JAV56586.1 APCN01003350 AAAB01008984 EAL38946.3 GEZM01077598 JAV63253.1 AB363979 BAG14261.1 GECZ01009869 JAS59900.1 KZ150278 PZC71645.1 GFTR01005802 JAW10624.1 GEBQ01017356 JAT22621.1 KK852672 KDR18777.1 GEBQ01022601 JAT17376.1 ATLV01018364 KE525231 KFB42999.1 GEBQ01016151 JAT23826.1 NWSH01004041 PCG65541.1 GEDC01011978 JAS25320.1 KU218697 ALX00077.1 ADMH02000599 ETN65690.1 GFDF01005349 JAV08735.1 AXCN02000734 PZC70661.1 AJWK01025808 AJWK01025809 AXCM01000044 GALX01005659 JAB62807.1 GFXV01000630 MBW12435.1 GFXV01005197 MBW17002.1
BABH01036387 BABH01036388 BABH01036389 BABH01036390 BABH01036391 AY672797 AAV91019.1 KQ460127 KPJ17583.1 GAIX01012077 JAA80483.1 NWSH01000257 PCG77964.1 ODYU01004095 SOQ43606.1 AY672798 AAV91020.1 RSAL01000491 RVE41548.1 KQ459249 KPJ02244.1 AF017664 AAB94558.1 KPJ02240.1 MH200930 AXS59140.1 AY672789 AAV91011.1 KPJ02243.1 AGBW02009991 OWR49389.1 KQ460130 KPJ17530.1 KZ150408 PZC70992.1 KZ150503 PZC70659.1 ODYU01002875 SOQ40903.1 NWSH01000265 PCG77882.1 KZ150484 PZC70714.1 KPJ17576.1 NWSH01000060 PCG80065.1 ODYU01002282 SOQ39557.1 AY672794 AAV91016.1 KZ150189 PZC72424.1 AGBW02010978 OWR47436.1 KZ150344 PZC71293.1 KZ150652 PZC70412.1 NWSH01001045 PCG72935.1 PCG72936.1 ODYU01002903 SOQ40975.1 ODYU01000940 SOQ36457.1 NWSH01002560 PCG67981.1 ODYU01006166 SOQ47783.1 KZ150324 PZC71401.1 GANO01003282 JAB56589.1 PZC71287.1 BABH01038780 NWSH01005248 PCG64337.1 NWSH01003268 PCG66648.1 ODYU01004086 SOQ43592.1 SOQ43594.1 DS231821 EDS44979.1 GFDL01005857 JAV29188.1 KZ150249 PZC71840.1 NWSH01000008 PCG80964.1 CH477825 EAT35883.2 NEVH01018378 PNF23527.1 UFQT01000075 UFQT01000299 SSX19399.1 SSX22964.1 KQ971311 EEZ99357.2 ODYU01003947 SOQ43326.1 GEZM01092351 JAV56586.1 APCN01003350 AAAB01008984 EAL38946.3 GEZM01077598 JAV63253.1 AB363979 BAG14261.1 GECZ01009869 JAS59900.1 KZ150278 PZC71645.1 GFTR01005802 JAW10624.1 GEBQ01017356 JAT22621.1 KK852672 KDR18777.1 GEBQ01022601 JAT17376.1 ATLV01018364 KE525231 KFB42999.1 GEBQ01016151 JAT23826.1 NWSH01004041 PCG65541.1 GEDC01011978 JAS25320.1 KU218697 ALX00077.1 ADMH02000599 ETN65690.1 GFDF01005349 JAV08735.1 AXCN02000734 PZC70661.1 AJWK01025808 AJWK01025809 AXCM01000044 GALX01005659 JAB62807.1 GFXV01000630 MBW12435.1 GFXV01005197 MBW17002.1
Proteomes
UP000005204
UP000053240
UP000218220
UP000283053
UP000053268
UP000007151
+ More
UP000002320 UP000008820 UP000235965 UP000007266 UP000076407 UP000075840 UP000075903 UP000007062 UP000075902 UP000075882 UP000076408 UP000027135 UP000075900 UP000030765 UP000075880 UP000075884 UP000069272 UP000000673 UP000075920 UP000075886 UP000092461 UP000075883
UP000002320 UP000008820 UP000235965 UP000007266 UP000076407 UP000075840 UP000075903 UP000007062 UP000075902 UP000075882 UP000076408 UP000027135 UP000075900 UP000030765 UP000075880 UP000075884 UP000069272 UP000000673 UP000075920 UP000075886 UP000092461 UP000075883
Interpro
Gene 3D
CDD
ProteinModelPortal
F5BZV1
H9JJA9
Q5MPB3
A0A0N1PHF7
S4NXP2
A0A2A4K236
+ More
A0A2H1VS51 Q5MPB2 A0A3S2NPZ4 A0A194Q9Z8 O44331 A0A194QB71 A0A346RAE0 Q5MPC1 A0A194Q9Q9 A0A212F6Q1 A0A0N0PDL6 A0A2W1B2N2 A0A2W1B0L2 A0A2H1VJ99 A0A2A4K2L1 A0A2W1B9D9 A0A0N1IHL2 A0A2A4K714 A0A2H1VFF0 Q5MPB6 A0A2W1BFG9 A0A212F107 A0A2W1BHZ8 A0A2W1AYZ6 A0A2A4JLI0 A0A2A4JMC3 A0A2H1VJG7 A0A2H1V6L6 A0A2A4J7N6 A0A2H1W424 A0A2W1B9E2 U5ESE3 A0A2W1BD72 H9JLA5 A0A2A4IWJ7 A0A2A4J4P7 A0A2H1VSY3 A0A2H1VTL8 B0W1A9 A0A1Q3FNX1 A0A2W1BGC1 A0A2A4KAM6 Q16NE9 A0A2J7Q4L6 A0A336LYA5 D6WCU1 A0A2H1VRC0 A0A182XUQ8 A0A1Y1KAU3 A0A182IIN2 A0A182VNZ6 Q5TNC5 A0A182THH3 A0A1Y1KPC1 B1B5K0 A0A182KNG1 A0A182Y982 A0A1B6GBU6 A0A2W1BCP3 A0A224XHV2 A0A1B6LG03 A0A067RHT4 A0A1B6L129 A0A182S507 A0A084VYF5 A0A182JES5 A0A1B6LJF7 A0A182NZ25 A0A2A4J0N6 A0A1B6DI71 A0A182FZY7 A0A0U4B511 W5JRY9 A0A1L8DQW0 A0A182WR24 A0A182Q673 A0A2W1B5J0 A0A1B0CS99 A0A182MQJ3 V5GMH2 A0A2H8TED5 A0A2H8TTU6
A0A2H1VS51 Q5MPB2 A0A3S2NPZ4 A0A194Q9Z8 O44331 A0A194QB71 A0A346RAE0 Q5MPC1 A0A194Q9Q9 A0A212F6Q1 A0A0N0PDL6 A0A2W1B2N2 A0A2W1B0L2 A0A2H1VJ99 A0A2A4K2L1 A0A2W1B9D9 A0A0N1IHL2 A0A2A4K714 A0A2H1VFF0 Q5MPB6 A0A2W1BFG9 A0A212F107 A0A2W1BHZ8 A0A2W1AYZ6 A0A2A4JLI0 A0A2A4JMC3 A0A2H1VJG7 A0A2H1V6L6 A0A2A4J7N6 A0A2H1W424 A0A2W1B9E2 U5ESE3 A0A2W1BD72 H9JLA5 A0A2A4IWJ7 A0A2A4J4P7 A0A2H1VSY3 A0A2H1VTL8 B0W1A9 A0A1Q3FNX1 A0A2W1BGC1 A0A2A4KAM6 Q16NE9 A0A2J7Q4L6 A0A336LYA5 D6WCU1 A0A2H1VRC0 A0A182XUQ8 A0A1Y1KAU3 A0A182IIN2 A0A182VNZ6 Q5TNC5 A0A182THH3 A0A1Y1KPC1 B1B5K0 A0A182KNG1 A0A182Y982 A0A1B6GBU6 A0A2W1BCP3 A0A224XHV2 A0A1B6LG03 A0A067RHT4 A0A1B6L129 A0A182S507 A0A084VYF5 A0A182JES5 A0A1B6LJF7 A0A182NZ25 A0A2A4J0N6 A0A1B6DI71 A0A182FZY7 A0A0U4B511 W5JRY9 A0A1L8DQW0 A0A182WR24 A0A182Q673 A0A2W1B5J0 A0A1B0CS99 A0A182MQJ3 V5GMH2 A0A2H8TED5 A0A2H8TTU6
PDB
2OLG
E-value=3.43986e-30,
Score=328
Ontologies
GO
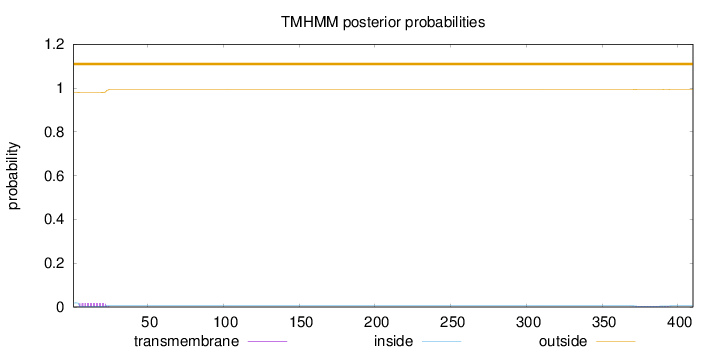
Topology
SignalP
Position: 1 - 19,
Likelihood: 0.998261
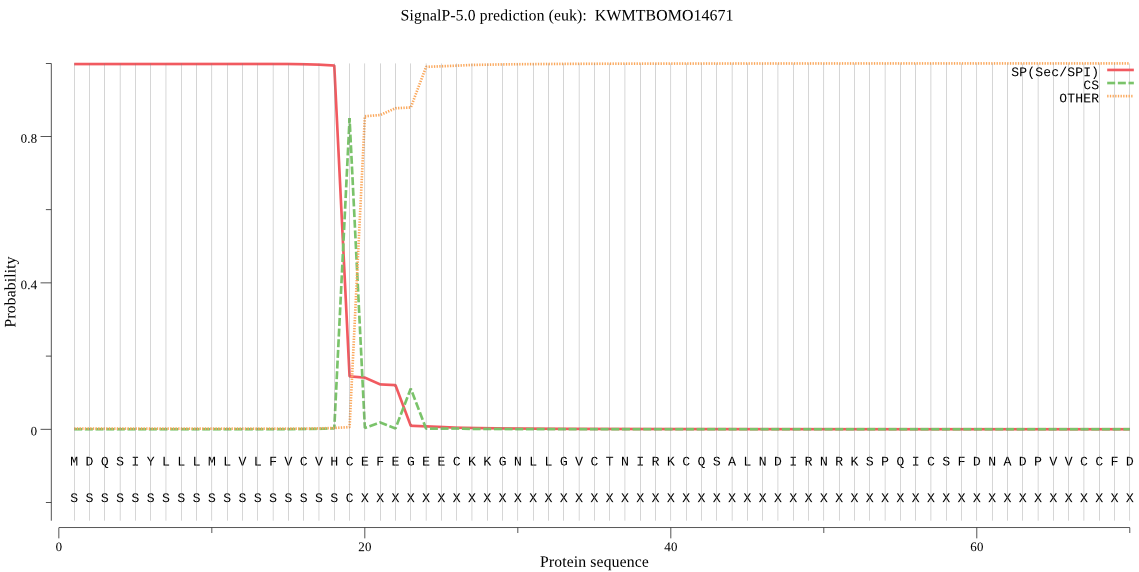
Length:
410
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.417810000000001
Exp number, first 60 AAs:
0.33347
Total prob of N-in:
0.01874
outside
1 - 410
Population Genetic Test Statistics
Pi
255.897749
Theta
158.088883
Tajima's D
3.303905
CLR
0.114536
CSRT
0.989200539973001
Interpretation
Uncertain
