Gene
KWMTBOMO14669
Pre Gene Modal
BGIBMGA009610
Annotation
PREDICTED:_uncharacterized_protein_LOC101736046_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.027 Nuclear Reliability : 1.475
Sequence
CDS
ATGCTGTATTATTTTTTTGCTTATAGAGATGGACGACCTCACAGCTCACCTGGTGCTAAGTGGTCACCGGTGCCCACAGACATCAAGAATACAAATGCACCCCCGGAACTAGTTTTGAAGATATCGTACATGGCCTTCGAACATGACACAGCGAATCTTCTCACGGAACACCACAAAACGTACTGGAACATATTAGCGGATAGACTGAATTATTTAGAGAAATTAAAGACCAAAGTTAGCATAACCAAATTAATGAACAAACTAACCGAAGACATATTGAAACTAATGGAGATATATGACACCGTTCAATTTTGTGTGAATGTTCTAAGTTATTTAGTTAAGAAACTGCACAGCCTGTACAATCAGAACGATTTTGTTGAACTAAACAAGCTTTATTTGAACTTGTTCGAGGGAATGTCCAAGAAAAGCGATTTGAATGCTTTCAATAGATTACCCGAGAAGGAATTGCTGGATGTGTACTTCAAGTTGAACGATTGCTTTTACGTCATCGCTGAGAACGCTTCGAAGAATGACTTTAGAGATGCGAAATTGGAAATCATAGTACGCTGCATCATCAGTTTGGTCGGTCAAGACAGAACTTTATTTAATTTGATACAAACATTCTATATGAATTCCTTCTGTTGTATATTAACTGAAAATATCGATGAAGTGTACGCTGAGAGGATATTAGAAAGTATATTAAGATCGTTTGAGGCAACCGAAAGACTCGGCTACAATAATGCATTGAACGTGACATATCCATTCTTGAATCAACTGTTAAGGTTGTTTATAGAGAAGCATGAAAGCGTTAATGTGAACACACAAGAGAAGGCACTGAAGTTGATCTTGTATCTCATGGGCAAGCTGGGGCACACGAAGCAGTTTCTAAAGTGTGAGAACTGTAAGTCGGCAACAGGTTGGCACGATGCCCTAAAGCTATCATTCCAAGTTAAGAATATACTGAGCGCATCCTTGAGCAGACAGTTAGATGTCAGGACCATCTTACCGATCATCTATCAGATCATAAAGGAGCAATACGCTGTATTGAATAGATTGATTCAGTTGAAATGTGTCAATGGCATGAAATTCTTTAGGAAGCTACAAACAGACGTTCACAATTTAGCAATTAATCTGAATAAAGAATCTTACAACGAACAATCTGTCAAACTTTTCAGTGTTTACTTAAAATATGAGATAGCCTATTATAGTAATGAAGCGGACTCGAAGAATATAAGCAGAGCTCTTTACAACAAGAGTATCTGTGAAATGGATTCGAAGATGTACGATGAAGCCCTCACGGATGCCTACCTCAGTTTGATTTTCAATTTGTCCGATAAACAAACTGACAAGCACATGAATTTGGTGATGGACATCAAGGCTAAGGCTGCGAAAGCCATCGACGACGAGGCTCTGCAGCTGCGGAGTGTGCTGGAAGCTTGCAAGGTGGCCGTTGAGAAAGAGCTTTACGGAAACATAAGAACTTTCTTTGTTGGTGTCAAATTTAGCGAGATCCTGCAGCACGAGTTTTCAATGTACGCCAAGCAGTGGCCCTCGATGGTTCCAGTGGCAGGCGTGTGGACCAGCCTGTGGGATCTGCTGCAGGGGCGGCACCCCTGGACGCACGCGGAACAAGAGAGCGTTTACAAGTGGGCCCTCTATGACGTCATGCTCCAGACCCCTGATCACGTGAGGAGTCTGCACTGCAAGCACTATCGGAGGGTCGTCAATATCGTCATCGACTACATGGATCGGAATGGGACTAAAGAAGTTGACGAGAATATCGTGTACGTGACGCTGCTGTTTCTGAAGTCCGAGATCGACTTGTGTGAGGCCAGCGAGGTGCACGGCTGGAGGGCCGGAGAGGCGAATTCAGATCCTGACAAGGTCCAATACACTAGGACCCTCAGGCAGGAGCACGACGCGACCAGACCGGCCCTGCGCGCGGTGCCGCTGTGGGGGGACCTCGTGCCCCACCTGGACCGAGGTGGGTTGAGACATGAGGTCGAGGACTCATTGCTGAGAGCGCCGCCCCTTGCGCGGGTGTCGCCTGAACGCACGACTTACTAA
Protein
MLYYFFAYRDGRPHSSPGAKWSPVPTDIKNTNAPPELVLKISYMAFEHDTANLLTEHHKTYWNILADRLNYLEKLKTKVSITKLMNKLTEDILKLMEIYDTVQFCVNVLSYLVKKLHSLYNQNDFVELNKLYLNLFEGMSKKSDLNAFNRLPEKELLDVYFKLNDCFYVIAENASKNDFRDAKLEIIVRCIISLVGQDRTLFNLIQTFYMNSFCCILTENIDEVYAERILESILRSFEATERLGYNNALNVTYPFLNQLLRLFIEKHESVNVNTQEKALKLILYLMGKLGHTKQFLKCENCKSATGWHDALKLSFQVKNILSASLSRQLDVRTILPIIYQIIKEQYAVLNRLIQLKCVNGMKFFRKLQTDVHNLAINLNKESYNEQSVKLFSVYLKYEIAYYSNEADSKNISRALYNKSICEMDSKMYDEALTDAYLSLIFNLSDKQTDKHMNLVMDIKAKAAKAIDDEALQLRSVLEACKVAVEKELYGNIRTFFVGVKFSEILQHEFSMYAKQWPSMVPVAGVWTSLWDLLQGRHPWTHAEQESVYKWALYDVMLQTPDHVRSLHCKHYRRVVNIVIDYMDRNGTKEVDENIVYVTLLFLKSEIDLCEASEVHGWRAGEANSDPDKVQYTRTLRQEHDATRPALRAVPLWGDLVPHLDRGGLRHEVEDSLLRAPPLARVSPERTTY
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
EMBL
Proteomes
PRIDE
Pfam
PF00089 Trypsin
Interpro
SUPFAM
SSF50494
SSF50494
CDD
ProteinModelPortal
Ontologies
GO
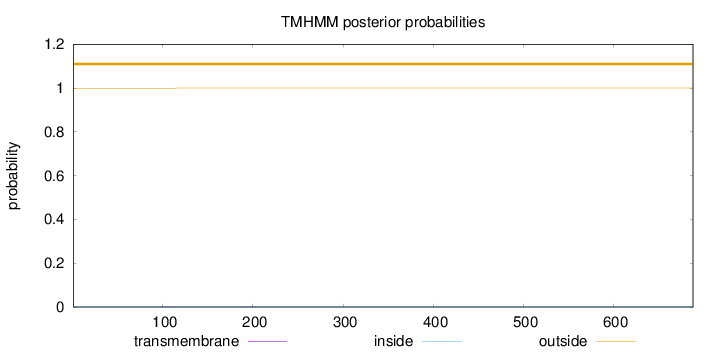
Topology
Length:
688
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00767999999999998
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00049
outside
1 - 688
Population Genetic Test Statistics
Pi
192.631067
Theta
154.623933
Tajima's D
0.936133
CLR
150.634951
CSRT
0.64306784660767
Interpretation
Uncertain
