Pre Gene Modal
BGIBMGA009608
Annotation
PREDICTED:_phosphatidate_phosphatase_PPAPDC1B_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.622
Sequence
CDS
ATGCCTCTCGTACGCTCGGCTAGTTTCAACTTGTTAATAGAAATCGTGTTAAGAATCGCTTTAATGTCAGCATTTTGTTACATGGAGACTCTGCCCCCTTTCAAGCGAGTAATTCACCCGGCGGACTTGGAGTCGCTTTACAAGTATCCCCGTGTGGATTCCTACGTTCCTGGGGGCACGTTATGGGCCATAGTGCTGTCGGTGCCTTGTGTACTGTCGATATTGGCTTGGGGTTGCTGCAACGACTGCAATGACGCCTTCGAGATACTACTAGCGTGGTCGTTAGCTCTAGGTCTGAACGGCGTTTTGACCGACGGAATAAAATTGATAGCAGGACGGCCGAGACCAGACTTCTTCTACAGATGCTTCCCCGACGGAGTGGAGACTCCGAAGCTGGTCTGCACTGGGGATCCGATGGTCGTGATGGACGGAAGGAAGTCTTTCCCGAGCGGTCACAGCAGCTTGTCGTTCTGCTCATTGGGCGTCGCTTCCGTTTGGCTCTGCGGGCGGCTGGGTACAACCTCGCGCCGGCGGGGCAGTGCAGCGAGGGTCCTGATCACCCTCTGCCCCCTGGTCGTGGCCGGCATAGTCGCTCTATCGCGCTCCTGCGATTACCATCATCACTGGGAAGATATCCTCGTGGGATCTTTACTTGGGTTCACGATTGCAGTTTTCTGCTACAGACAATACTACAATCCTCTGTCTTCAGACCTGGCAGGGGTTCCGTATATAGTCTCCAATAACATGGCCAAGTACGGAAAAGAAATAAGTCCGACGAAAGATTTAAAGGAAGTCGAATCTACTCCGCTTTTGGGGAAGAAAGATGATAAATGGATCTGA
Protein
MPLVRSASFNLLIEIVLRIALMSAFCYMETLPPFKRVIHPADLESLYKYPRVDSYVPGGTLWAIVLSVPCVLSILAWGCCNDCNDAFEILLAWSLALGLNGVLTDGIKLIAGRPRPDFFYRCFPDGVETPKLVCTGDPMVVMDGRKSFPSGHSSLSFCSLGVASVWLCGRLGTTSRRRGSAARVLITLCPLVVAGIVALSRSCDYHHHWEDILVGSLLGFTIAVFCYRQYYNPLSSDLAGVPYIVSNNMAKYGKEISPTKDLKEVESTPLLGKKDDKWI
Summary
Uniprot
H9JJA7
A0A2A4K976
A0A2H1VHS2
A0A194QA04
A0A212END8
A0A0P5FIM5
+ More
A0A164ZVA3 A0A0P4Y150 A0A2W1BRT5 A0A2A4JXK8 E9HAW1 A0A0P6F278 A0A0P5TCE8 A0A088AF86 A0A2A3EQY0 A0A0L7L150 A0A0P6BBS7 A0A0P6ETW1 A0A310S9Q7 A0A182PQV9 A0A2P6KCC9 A0A182JP82 A0A195BEL6 A0A195FHZ3 A0A182RXK9 A0A182VR20 A0A026WJX1 A0A151IYI3 A0A182YQM1 A0A2M4AA62 A0A182V2J9 Q7PYH4 G3H1J0 A0A023EPA5 A0A084VS39 A0A182MDC0 A0A182T9E9 A0A1S2ZBE7 A0A2M4BRX6 G1PG55 A0A2M3Z3U8 A0A182GCJ8 A0A2M4BRP5 A0A2M3Z389 A0A2M4BRT0 S7MD21 A0A2M4BRQ2 A0A182F891 A0A2M4BRS4 E0VG45 A0A1J1IZN8 A0A2H1V7N7 A0A182I0B7 Q17FK0 A0A182JKV8 W5JDC1 R4G9L0 H9G5X7 A0A232F757 K7INY7 A0A023EQH7 E2BNH2 A0A151IKX1 A0A182XBH7 A0A154PSV6 A0A0B8RWT0 A0A182U6L1 A0A182QI68 A0A3P8WUY8 A0A2J7QSN0 I3KAH8 A0A3B4FN78 A0A3Q4HT90 A0A3P8PIC3 A0A3P9BS93 A0A3P9K7N9 A0A3Q2VKM1 A0A2K6QGD4 H2LBZ7 F6VCP3 F6YGV2 A0A3Q2XJ21 A0A093BRK8 A0A1A7XY65 A0A0P6J654 A0A2L2Y7N3 A0A110AI45 A0A151WEL3 A0A3B3CFK8 A0A3Q3A0F5 R4GC42 A0A093HQP3 A0A2J8PC03 A0A2J8W6L4 A0A0A0MSZ0 A0A182N615 A0A2K6CQ07 A0A2K5TNH1 A0A2K5ZPF5 A0A2R9C2Y2
A0A164ZVA3 A0A0P4Y150 A0A2W1BRT5 A0A2A4JXK8 E9HAW1 A0A0P6F278 A0A0P5TCE8 A0A088AF86 A0A2A3EQY0 A0A0L7L150 A0A0P6BBS7 A0A0P6ETW1 A0A310S9Q7 A0A182PQV9 A0A2P6KCC9 A0A182JP82 A0A195BEL6 A0A195FHZ3 A0A182RXK9 A0A182VR20 A0A026WJX1 A0A151IYI3 A0A182YQM1 A0A2M4AA62 A0A182V2J9 Q7PYH4 G3H1J0 A0A023EPA5 A0A084VS39 A0A182MDC0 A0A182T9E9 A0A1S2ZBE7 A0A2M4BRX6 G1PG55 A0A2M3Z3U8 A0A182GCJ8 A0A2M4BRP5 A0A2M3Z389 A0A2M4BRT0 S7MD21 A0A2M4BRQ2 A0A182F891 A0A2M4BRS4 E0VG45 A0A1J1IZN8 A0A2H1V7N7 A0A182I0B7 Q17FK0 A0A182JKV8 W5JDC1 R4G9L0 H9G5X7 A0A232F757 K7INY7 A0A023EQH7 E2BNH2 A0A151IKX1 A0A182XBH7 A0A154PSV6 A0A0B8RWT0 A0A182U6L1 A0A182QI68 A0A3P8WUY8 A0A2J7QSN0 I3KAH8 A0A3B4FN78 A0A3Q4HT90 A0A3P8PIC3 A0A3P9BS93 A0A3P9K7N9 A0A3Q2VKM1 A0A2K6QGD4 H2LBZ7 F6VCP3 F6YGV2 A0A3Q2XJ21 A0A093BRK8 A0A1A7XY65 A0A0P6J654 A0A2L2Y7N3 A0A110AI45 A0A151WEL3 A0A3B3CFK8 A0A3Q3A0F5 R4GC42 A0A093HQP3 A0A2J8PC03 A0A2J8W6L4 A0A0A0MSZ0 A0A182N615 A0A2K6CQ07 A0A2K5TNH1 A0A2K5ZPF5 A0A2R9C2Y2
Pubmed
19121390
26354079
22118469
28756777
21292972
26227816
+ More
24508170 30249741 25244985 12364791 14747013 17210077 21804562 24945155 24438588 21993624 26483478 20566863 17510324 20920257 23761445 21881562 28648823 20075255 20798317 25476704 24487278 25186727 17554307 25362486 19892987 25243066 26561354 29451363 15164054 22722832
24508170 30249741 25244985 12364791 14747013 17210077 21804562 24945155 24438588 21993624 26483478 20566863 17510324 20920257 23761445 21881562 28648823 20075255 20798317 25476704 24487278 25186727 17554307 25362486 19892987 25243066 26561354 29451363 15164054 22722832
EMBL
BABH01036368
BABH01036369
BABH01036370
BABH01036371
NWSH01000015
PCG80825.1
+ More
ODYU01002597 SOQ40311.1 KQ459249 KPJ02249.1 AGBW02013680 OWR42989.1 GDIQ01256621 JAJ95103.1 LRGB01000687 KZS16817.1 GDIP01233986 JAI89415.1 KZ149961 PZC76325.1 NWSH01000478 PCG76122.1 GL732613 EFX71150.1 GDIQ01054133 JAN40604.1 GDIP01129443 JAL74271.1 KZ288199 PBC33626.1 JTDY01003791 KOB69004.1 GDIP01016843 JAM86872.1 GDIQ01070970 JAN23767.1 KQ767877 OAD53271.1 MWRG01016230 PRD23964.1 KQ976509 KYM82654.1 KQ981606 KYN39609.1 KK107207 QOIP01000002 EZA55409.1 RLU25765.1 KQ980754 KYN13620.1 GGFK01004373 MBW37694.1 AAAB01008987 EAA01268.5 JH000106 EGV94664.1 GAPW01002778 JAC10820.1 ATLV01015797 KE525036 KFB40783.1 AXCM01006008 GGFJ01006613 MBW55754.1 AAPE02012468 GGFM01002445 MBW23196.1 JXUM01159949 JXUM01159950 KQ573526 KXJ67924.1 GGFJ01006614 MBW55755.1 GGFM01002236 MBW22987.1 GGFJ01006645 MBW55786.1 KE161075 EPQ01551.1 GGFJ01006615 MBW55756.1 GGFJ01006644 MBW55785.1 DS235131 EEB12351.1 CVRI01000065 CRL05669.1 ODYU01000918 SOQ36402.1 APCN01002059 CH477270 EAT45343.1 ADMH02001488 ETN62347.1 AAWZ02027246 NNAY01000833 OXU26273.1 GAPW01002764 JAC10834.1 GL449414 EFN82751.1 KQ977151 KYN05260.1 KQ435180 KZC14991.1 GBSH01002034 JAG66992.1 AXCN02000193 NEVH01011876 PNF31588.1 AERX01025333 GAMS01008859 GAMR01010613 GAMP01006053 JAB14277.1 JAB23319.1 JAB46702.1 KL227329 KFV03446.1 HADW01006038 HADX01000473 SBP22705.1 GEBF01003900 JAN99732.1 IAAA01018716 IAAA01018717 IAAA01018718 IAAA01018719 IAAA01018720 LAA04017.1 KT959203 AMB27083.1 KQ983238 KYQ46280.1 KL206697 KFV84993.1 NBAG03000216 PNI81545.1 NDHI03003398 PNJ65425.1 AC023282 AC073587 AL157782 AQIA01072795 AQIA01072796 AJFE02090426 AJFE02090427 AJFE02090428 AJFE02090429
ODYU01002597 SOQ40311.1 KQ459249 KPJ02249.1 AGBW02013680 OWR42989.1 GDIQ01256621 JAJ95103.1 LRGB01000687 KZS16817.1 GDIP01233986 JAI89415.1 KZ149961 PZC76325.1 NWSH01000478 PCG76122.1 GL732613 EFX71150.1 GDIQ01054133 JAN40604.1 GDIP01129443 JAL74271.1 KZ288199 PBC33626.1 JTDY01003791 KOB69004.1 GDIP01016843 JAM86872.1 GDIQ01070970 JAN23767.1 KQ767877 OAD53271.1 MWRG01016230 PRD23964.1 KQ976509 KYM82654.1 KQ981606 KYN39609.1 KK107207 QOIP01000002 EZA55409.1 RLU25765.1 KQ980754 KYN13620.1 GGFK01004373 MBW37694.1 AAAB01008987 EAA01268.5 JH000106 EGV94664.1 GAPW01002778 JAC10820.1 ATLV01015797 KE525036 KFB40783.1 AXCM01006008 GGFJ01006613 MBW55754.1 AAPE02012468 GGFM01002445 MBW23196.1 JXUM01159949 JXUM01159950 KQ573526 KXJ67924.1 GGFJ01006614 MBW55755.1 GGFM01002236 MBW22987.1 GGFJ01006645 MBW55786.1 KE161075 EPQ01551.1 GGFJ01006615 MBW55756.1 GGFJ01006644 MBW55785.1 DS235131 EEB12351.1 CVRI01000065 CRL05669.1 ODYU01000918 SOQ36402.1 APCN01002059 CH477270 EAT45343.1 ADMH02001488 ETN62347.1 AAWZ02027246 NNAY01000833 OXU26273.1 GAPW01002764 JAC10834.1 GL449414 EFN82751.1 KQ977151 KYN05260.1 KQ435180 KZC14991.1 GBSH01002034 JAG66992.1 AXCN02000193 NEVH01011876 PNF31588.1 AERX01025333 GAMS01008859 GAMR01010613 GAMP01006053 JAB14277.1 JAB23319.1 JAB46702.1 KL227329 KFV03446.1 HADW01006038 HADX01000473 SBP22705.1 GEBF01003900 JAN99732.1 IAAA01018716 IAAA01018717 IAAA01018718 IAAA01018719 IAAA01018720 LAA04017.1 KT959203 AMB27083.1 KQ983238 KYQ46280.1 KL206697 KFV84993.1 NBAG03000216 PNI81545.1 NDHI03003398 PNJ65425.1 AC023282 AC073587 AL157782 AQIA01072795 AQIA01072796 AJFE02090426 AJFE02090427 AJFE02090428 AJFE02090429
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000076858
UP000000305
+ More
UP000005203 UP000242457 UP000037510 UP000075885 UP000075881 UP000078540 UP000078541 UP000075900 UP000075920 UP000053097 UP000279307 UP000078492 UP000076408 UP000075903 UP000007062 UP000001075 UP000030765 UP000075883 UP000075901 UP000079721 UP000001074 UP000069940 UP000249989 UP000069272 UP000009046 UP000183832 UP000075840 UP000008820 UP000075880 UP000000673 UP000001646 UP000215335 UP000002358 UP000008237 UP000078542 UP000076407 UP000076502 UP000075902 UP000075886 UP000265120 UP000235965 UP000005207 UP000261460 UP000261580 UP000265100 UP000265160 UP000265180 UP000265200 UP000264840 UP000233200 UP000001038 UP000002281 UP000008225 UP000264820 UP000075809 UP000261560 UP000264800 UP000053584 UP000005640 UP000075884 UP000233120 UP000233100 UP000233140 UP000240080
UP000005203 UP000242457 UP000037510 UP000075885 UP000075881 UP000078540 UP000078541 UP000075900 UP000075920 UP000053097 UP000279307 UP000078492 UP000076408 UP000075903 UP000007062 UP000001075 UP000030765 UP000075883 UP000075901 UP000079721 UP000001074 UP000069940 UP000249989 UP000069272 UP000009046 UP000183832 UP000075840 UP000008820 UP000075880 UP000000673 UP000001646 UP000215335 UP000002358 UP000008237 UP000078542 UP000076407 UP000076502 UP000075902 UP000075886 UP000265120 UP000235965 UP000005207 UP000261460 UP000261580 UP000265100 UP000265160 UP000265180 UP000265200 UP000264840 UP000233200 UP000001038 UP000002281 UP000008225 UP000264820 UP000075809 UP000261560 UP000264800 UP000053584 UP000005640 UP000075884 UP000233120 UP000233100 UP000233140 UP000240080
PRIDE
Pfam
PF01569 PAP2
Interpro
SUPFAM
SSF48317
SSF48317
ProteinModelPortal
H9JJA7
A0A2A4K976
A0A2H1VHS2
A0A194QA04
A0A212END8
A0A0P5FIM5
+ More
A0A164ZVA3 A0A0P4Y150 A0A2W1BRT5 A0A2A4JXK8 E9HAW1 A0A0P6F278 A0A0P5TCE8 A0A088AF86 A0A2A3EQY0 A0A0L7L150 A0A0P6BBS7 A0A0P6ETW1 A0A310S9Q7 A0A182PQV9 A0A2P6KCC9 A0A182JP82 A0A195BEL6 A0A195FHZ3 A0A182RXK9 A0A182VR20 A0A026WJX1 A0A151IYI3 A0A182YQM1 A0A2M4AA62 A0A182V2J9 Q7PYH4 G3H1J0 A0A023EPA5 A0A084VS39 A0A182MDC0 A0A182T9E9 A0A1S2ZBE7 A0A2M4BRX6 G1PG55 A0A2M3Z3U8 A0A182GCJ8 A0A2M4BRP5 A0A2M3Z389 A0A2M4BRT0 S7MD21 A0A2M4BRQ2 A0A182F891 A0A2M4BRS4 E0VG45 A0A1J1IZN8 A0A2H1V7N7 A0A182I0B7 Q17FK0 A0A182JKV8 W5JDC1 R4G9L0 H9G5X7 A0A232F757 K7INY7 A0A023EQH7 E2BNH2 A0A151IKX1 A0A182XBH7 A0A154PSV6 A0A0B8RWT0 A0A182U6L1 A0A182QI68 A0A3P8WUY8 A0A2J7QSN0 I3KAH8 A0A3B4FN78 A0A3Q4HT90 A0A3P8PIC3 A0A3P9BS93 A0A3P9K7N9 A0A3Q2VKM1 A0A2K6QGD4 H2LBZ7 F6VCP3 F6YGV2 A0A3Q2XJ21 A0A093BRK8 A0A1A7XY65 A0A0P6J654 A0A2L2Y7N3 A0A110AI45 A0A151WEL3 A0A3B3CFK8 A0A3Q3A0F5 R4GC42 A0A093HQP3 A0A2J8PC03 A0A2J8W6L4 A0A0A0MSZ0 A0A182N615 A0A2K6CQ07 A0A2K5TNH1 A0A2K5ZPF5 A0A2R9C2Y2
A0A164ZVA3 A0A0P4Y150 A0A2W1BRT5 A0A2A4JXK8 E9HAW1 A0A0P6F278 A0A0P5TCE8 A0A088AF86 A0A2A3EQY0 A0A0L7L150 A0A0P6BBS7 A0A0P6ETW1 A0A310S9Q7 A0A182PQV9 A0A2P6KCC9 A0A182JP82 A0A195BEL6 A0A195FHZ3 A0A182RXK9 A0A182VR20 A0A026WJX1 A0A151IYI3 A0A182YQM1 A0A2M4AA62 A0A182V2J9 Q7PYH4 G3H1J0 A0A023EPA5 A0A084VS39 A0A182MDC0 A0A182T9E9 A0A1S2ZBE7 A0A2M4BRX6 G1PG55 A0A2M3Z3U8 A0A182GCJ8 A0A2M4BRP5 A0A2M3Z389 A0A2M4BRT0 S7MD21 A0A2M4BRQ2 A0A182F891 A0A2M4BRS4 E0VG45 A0A1J1IZN8 A0A2H1V7N7 A0A182I0B7 Q17FK0 A0A182JKV8 W5JDC1 R4G9L0 H9G5X7 A0A232F757 K7INY7 A0A023EQH7 E2BNH2 A0A151IKX1 A0A182XBH7 A0A154PSV6 A0A0B8RWT0 A0A182U6L1 A0A182QI68 A0A3P8WUY8 A0A2J7QSN0 I3KAH8 A0A3B4FN78 A0A3Q4HT90 A0A3P8PIC3 A0A3P9BS93 A0A3P9K7N9 A0A3Q2VKM1 A0A2K6QGD4 H2LBZ7 F6VCP3 F6YGV2 A0A3Q2XJ21 A0A093BRK8 A0A1A7XY65 A0A0P6J654 A0A2L2Y7N3 A0A110AI45 A0A151WEL3 A0A3B3CFK8 A0A3Q3A0F5 R4GC42 A0A093HQP3 A0A2J8PC03 A0A2J8W6L4 A0A0A0MSZ0 A0A182N615 A0A2K6CQ07 A0A2K5TNH1 A0A2K5ZPF5 A0A2R9C2Y2
Ontologies
PATHWAY
GO
PANTHER
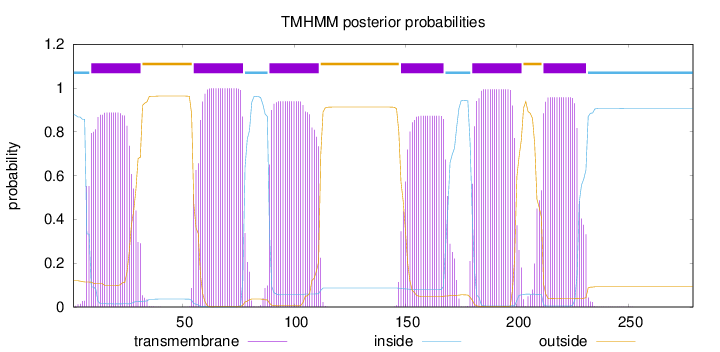
Topology
Length:
279
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
118.79613
Exp number, first 60 AAs:
23.59784
Total prob of N-in:
0.88007
POSSIBLE N-term signal
sequence
inside
1 - 8
TMhelix
9 - 31
outside
32 - 54
TMhelix
55 - 77
inside
78 - 88
TMhelix
89 - 111
outside
112 - 147
TMhelix
148 - 167
inside
168 - 179
TMhelix
180 - 202
outside
203 - 211
TMhelix
212 - 231
inside
232 - 279
Population Genetic Test Statistics
Pi
291.835724
Theta
186.852066
Tajima's D
2.063613
CLR
0.224782
CSRT
0.887755612219389
Interpretation
Uncertain
